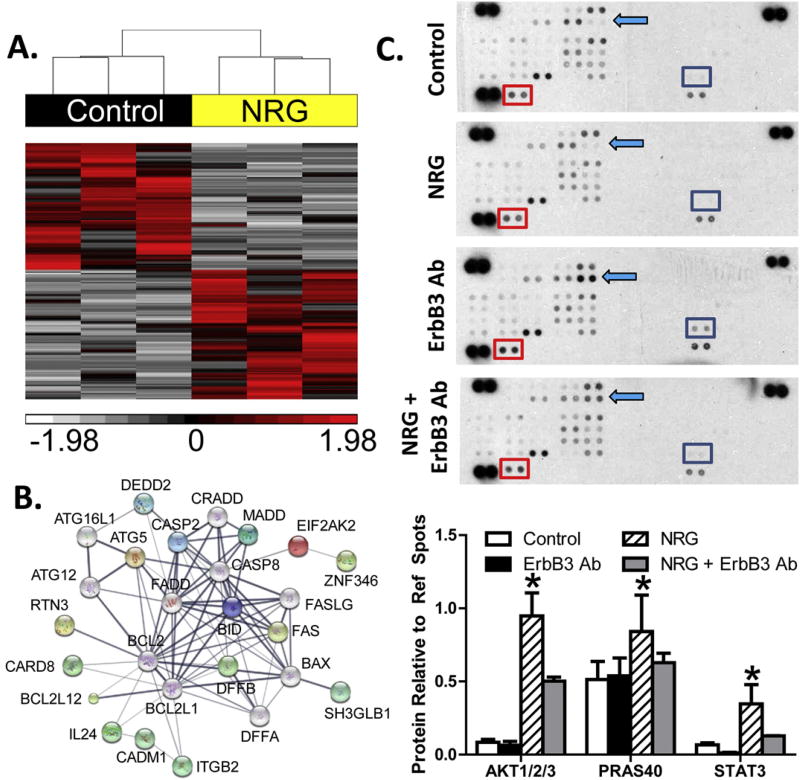
Fig. 2.
RNA sequencing and protein array results for neuregulin-treated fibroblasts. A, hierarchical clustering of RNA transcript RPKM (Reads Per Kilobase per Million mapped reads) for differential genes between untreated and NRG-1β-treated NHCV fibroblasts. The cluster was produced using Partek Genomics Suite with average linkage and Euclidian distance measures. Bright red, white, and black indicate the highest, lowest, and median normalized reads, respectively. Vertical dendrograms represent the individual donor fibroblasts, of which there are three replicates for each group. B, differentially expressed genes associated with cell death formed a functional protein interaction network when analyzed using STRING software code and libraries, which are freely available online (string-db.org). C, representative immunoblots show differentially expressed phosphorylated AKT pathway proteins in NHCV fibroblasts that were untreated or treated for 30 min with NRG-1β, with and without pre-treatment with MAB3481, an ErbB3 blocking antibody. Each protein is represented on the array in duplicate, and each array includes three positive references (top left, top right, and bottom left) as well as negative reference spots (bottom right). Colored squares indicate significantly differential proteins. D, graphical representation of differentially expressed proteins identified using immunoblot arrays. Data includes densitometry analysis results for four replicate experiments (n = 4, *p < 0.05 for NRG-1β-treated cells, when compared to the various control treatments).