Fig. 1.
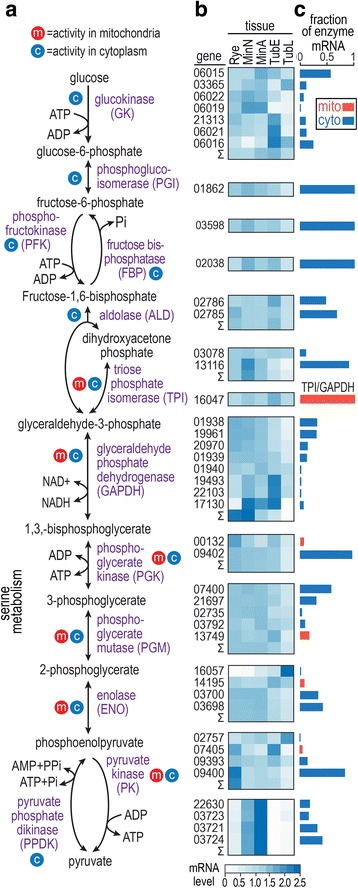
Glycolysis in Phytophthora infestans. a, Overview. Metabolites are indicated by black text and enzymes are indicated by purple text. Enzymes having mitochondrial or cytoplasmic forms are marked by red and blue circles, respectively. b, mRNA levels during growth on complex media (rye grain media), minimal media with glucose or amino acids as the main carbon source (MinA, MinN), and potato tubers at 1.5 and 4 days post-infection (TubE, TubL). Data are from RNA-seq analysis and are shown as per gene-normalized CPM values after TMM normalization by edgeR. Heatmaps are to the right of the corresponding enzymes in panel a. The five-digit numbers on the left side of the heatmap are the gene names, trimmed of the “PITG_” prefix. For enzyme activities produced by more than one gene, the sum of CPM values in each tissue type is represented by the row labeled Σ. c, Contribution of each gene to the total transcript pool for each enzyme, based on RPKM values. For example, the bar for glucokinase gene 06015 equals 0.54, which means that its transcripts represent 54% of all mRNAs encoding glucokinase. Mitochondrial and cytoplasmic forms of each enzyme are represented by red and blue bars, respectively
