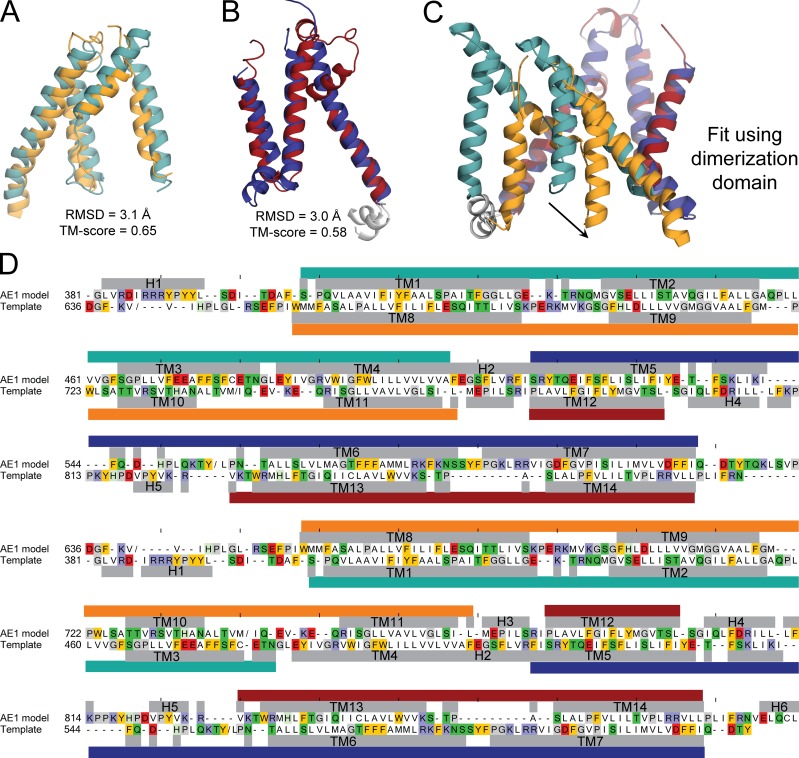
Figure 2.
Asymmetry in the structural repeats forming the membrane domain of AE1, and sequence alignment used to construct a repeat-swapped model. (A and B) Structural similarity between TM1–TM4 and TM8–TM11 in the transport domain (A) and TM5–TM7 and TM12–TM14 in the dimerization domain (B), compared using TMalign. The corresponding RMSD and TM-score values show that the difference between repeats (Fig. 1 C) is due to the reorientation of two units within each repeat. (A) The transport-domain segments comprise residues 381–508 and 636–775, of which 111 residues are aligned. (B) The dimerization-domain segments comprise residues 509–635 and 776–880, of which 88 residues are aligned. (C) Asymmetry of the two structural repeats. (D) Sequence alignment between AE1 model and the template, which is the x-ray crystal structure with the order of the repeats exchanged to B then A; the sequence identity is 8.3%. Residues with helical secondary structure according to DSSP are indicated with gray bars and labeled by helix or TM segment. Repeat A is indicated with teal and dark blue bars, and repeat B is indicated with orange and deep red bars (transport and dimerization domains, respectively). “/” indicates a break in the sequence if the segment was not modeled or was not present in the template structure (residues Y553 to L467, V640 to V649, and M741 to I753). Ticks above the model sequence are located every 10 positions for reference. Residues are colored if they are aromatic (gold), basic (indigo), acidic (red), glycine or proline (gray), or polar (green). Residues ELQCL in H6 were modeled using the same element from the crystal structure as a template after applying the transformation matrix that superposes the two repeats.