Pierce et al. describe a pediatric patient with a fatal systemic capillary leak syndrome (Clarkson’s disease). They identify a point mutation in p190BRhoGAP and show that patient-derived microvascular endothelial cells show prolonged activation RhoB that correlates with impaired barrier recovery after treatment with TNF compared with control cultures.
Abstract
We describe a fatal case of pediatric systemic capillary leak (Clarkson’s disease) associated with a point mutation in p190BRhoGAP. Dermal microvascular endothelial cells (ECs) isolated from this patient form monolayers with similar levels and distribution of junctional proteins and transendothelial electrical resistance compared with normal human dermal microvascular ECs. However, patient-derived ECs demonstrate a greater increase in permeability and impaired recovery of barrier function in response to tumor necrosis factor (TNF) compared with normal donor EC cultures. TNF transiently activates RhoB in ECs coincident with developing leak, and inactivation of RhoB correlates with barrier recovery. The mutation in p190BRhoGAP impairs RhoB inactivation, and the mutant phenotype of patient-derived ECs is replicated by siRNA knockdown of p190BRhoGAP in normal ECs. These data suggest a previously unknown function for p190BRhoGAP in control of capillary EC barrier function that may also be important in acquired systemic capillary leak associated with critical illness in humans.
Introduction
Endothelial cells (ECs) actively regulate blood flow and fluidity, immune cell quiescence and recruitment, as well as blood vessel permeability. In the context of systemic inflammation, pathological changes in blood vessel permeability, so called capillary leak, increase mortality via intravascular volume depletion, hemoconcentration, and tissue edema. The cellular mechanisms by which capillaries become leaky in human disease are not known. We present investigations into a congenital dysregulation of vascular permeability that provides insight into the mechanisms of capillary leak in human disease.
Idiopathic systemic capillary leak syndrome (SCLS, also known as Clarkson’s disease) is a rare disorder characterized by acute, recurrent episodes of vascular leak that manifest as severe hypotension, hemoconcentration, and hypoalbuminemia with generalized edema in the absence of overt inflammation or alternate etiology (Clarkson et al., 1960). Episodes may vary in frequency from weekly to several episodes per decade and range in severity from mild to lethal. More than 80% of adult patients with SCLS will have an associated monoclonal gammopathy of underdetermined significance. The 5-yr mortality rate in adults is estimated to be >75% (Xie et al., 2014). More than 300 cases have been described in adults, but only 18 have been described in children (Hsu et al., 2015). Pediatric SCLS occurs at a younger age without immune cell dyscrasias and is more likely to be inherited rather than acquired. Capillary leak from SCLS is presumed to result from the disruption of EC tight junctions (TJs) and is often more rapidly developing and severe than the capillary leak that occurs with systemic inflammation (Xie et al., 2014). Capillary leak in SCLS also occurs in the setting of minimal or absent systemic inflammation. The inciting events of the dramatic EC permeability changes in pediatric SCLS are unknown, as are the cellular mechanism of capillary leak. Both dysregulated mediator production with normal vascular cell function (Kinoshita et al., 2010; Xie et al., 2013) and intrinsic dysfunction of EC response to systemic signaling (Assaly et al., 2001; Xie et al., 2012) have been proposed. Here we present a patient with SCLS and increased capillary leak due to increased and sustained RhoB activity associated with a deleterious mutation in the regulatory protein p190BRhoGAP.
Results and discussion
Case presentation
A 12-yr-old boy was diagnosed with SCLS at 10 yr of age and had several mild and three severe episodes (requiring intensive care) over the course of his life starting at age 7 yr. He had no other significant medical history. He was seen in the pediatric emergency department and discharged the previous evening for nonlocalized abdominal pain that self-resolved. Hours after discharge, he was found unresponsive at home in cardiac arrest. Cardiopulmonary resuscitation was initiated, and he was transported to our emergency department where he was resuscitated with return of spontaneous circulation. At that time, he was noted to have dilated and nonreactive pupils. His initial hematocrit was 48%, 20% above his baseline, and albumin was <1 g/dL (below laboratory detection limit), both hallmarks of a severe SCLS episode. During resuscitation in the first 24 h, he required 4.5 liters of crystalloid and 4 liters of colloid volume resuscitation as well as infusions of dopamine, norepinephrine, and epinephrine to maintain age-appropriate mean arterial blood pressure. He developed pediatric acute respiratory distress syndrome with characteristic chest radiograph findings consistent with severe pulmonary edema and hemorrhage (Fig. 1 A). Laboratory studies revealed evidence of disseminated intravascular coagulation, acute renal failure, rhabdomyolysis, acute liver injury, cardiac ischemia, and severe anoxic brain injury. On the sixth hospital day, life-sustaining therapies were withdrawn, and the patient passed away.
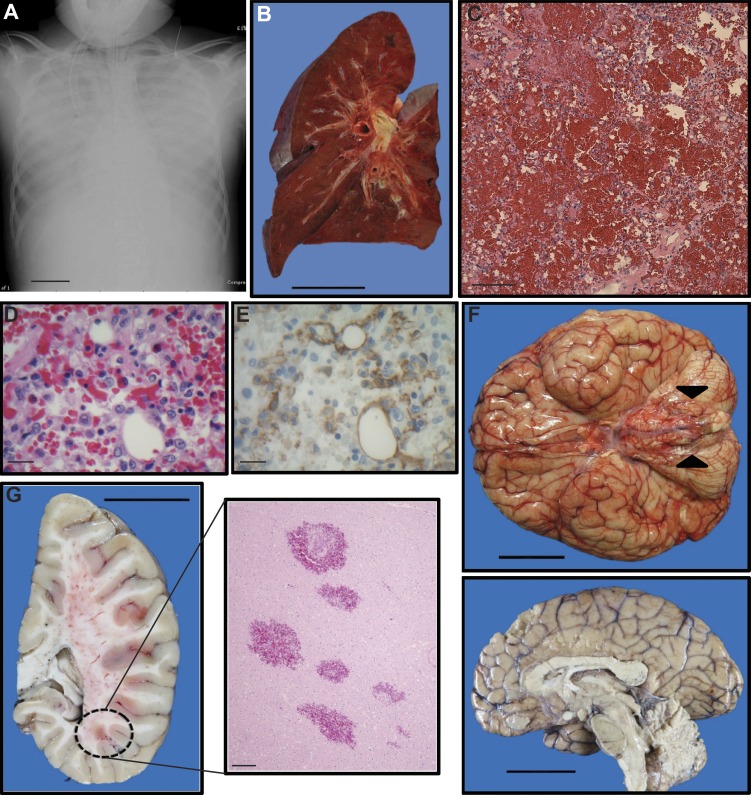
Figure 1.
Radiographical premortem and autopsy findings. (A) Chest x-ray during severe capillary leak indicating diffuse severe pulmonary edema. Bar, 6 cm. (B) Cross-sectional view of the left lung (formalin-inflated). Note that the lung is trilobed, an incidental finding. The fleshy appearance of the pulmonary parenchyma is conferred by the presence of intra-alveolar hemorrhage. Bar, 5 cm. (C) Representative view of pulmonary parenchyma with erythrocytes filling alveolar spaces (H&E stained). Bar, 100 µm. (D and E) High magnification view of pulmonary capillaries (H&E stained) with paired depictions of the same vessels illustrating capillary ECs (anti-CD31 antibody with brown chromogen via immunohistochemistry; note that nearby leukocytes also stain positively, an expected finding). Bars, 20 µm. (F) Inferior view of the brain (fresh). Edema and bilateral tonsillar herniation are evident (arrowheads), and the sagittal view of brain hemisection (formalin-fixed) depicts edema and ventricular narrowing. Bars, 5 cm. (G) Coronal section of sagittally divided brain hemisection (formalin-fixed) and corresponding histological section (H&E stained) demonstrating multiple intraparenchymal hemorrhages. Bars: (left) 5 cm; (right) 100 µm.
Autopsy analysis
Autopsy findings were consistent with the patient’s terminal clinical course. Although not an obese child, the patient’s postmortem weight was in the 93rd percentile for age and gender, despite a corresponding height in the 27th percentile, attributable to fluid retention, although generalized edema was only minimally evident. The extremities were devoid of swelling, with mild edema identified only in the tongue, lips, and scrotum. Notably, both ascites (100 ml) and a right pleural effusion (140 ml) were appreciated on autopsy. The most significant findings of the postmortem examination pertained to the lungs and brain. The former exhibited pulmonary hemorrhage bilaterally with concomitant evidence of diffuse alveolar damage, aspiration pneumonia, and pediatric acute respiratory distress syndrome (Fig. 1, B–E). Review of the brain demonstrated massive, diffuse cerebral edema with evidence of global hypoxic-ischemic encephalopathy as well as bilateral uncal and tonsillar herniation (Fig. 1, F and G). Additional autopsy findings (not depicted) included congestive hepatopathy, acute tubular necrosis in the kidney, hypertrophy of the cardiac left ventricular wall, and an incidentally identified trilobed left lung. Bone marrow examination revealed <5% plasma cells with normal kappa-lambda balance inconsistent with monoclonal gammopathy of undetermined significance, commonly associated with the adult form of SCLS. Other organ systems were without significant gross and histological abnormality. There was no evidence of myocardial infarction, infection, stroke, gastrointestinal bleed, or pulmonary embolism that could explain a severe episode of capillary leak. Thus, although the autopsy findings were consistent with the clinical course, they did not specifically identify the etiology of SCLS nor indicate a specific precipitating event for this terminal episode.
Dermal microvascular ECs isolated from the SCLS patient display normal junctional structures and G-protein receptor–induced responses but altered TNF responses
Human dermal microvascular ECs (HDMECs) isolated from a skin sample obtained at autopsy of the patient (SCLS HDMECs) demonstrated normal growth and morphology compared with HDMECs isolated from healthy donor patients (normal donor HDMECs). In the course of these studies, the SCLS HDMECs were compared with three to six different normal donor HDMEC isolates in each assay, although the donors were not necessarily age or gender matched. SCLS HDMECs form high-resistance barriers comparable to normal donor HDMECs (Fig. 2 A), previously shown to depend on TJs (Kluger et al., 2013). The ability of HDMECs to spontaneously form TJs, a property not shared by more commonly used cultured human umbilical vein ECs, makes them a good model for the study of capillary leak, which depends on TJ disruption. Consistent with high-resistance barriers, both SCLS and normal donor HDMECs display similar amounts and distribution of claudin-5, a specific organizing protein for HDMEC TJs (Kluger et al., 2013), by immunofluorescence microscopy (Fig. 2 B, left) and have similar ultrastructural appearance by electron microscopy (Fig. 2 D, top) under standard culture conditions. Specifically, immunofluorescence microscopy showed that claudin-5 was concentrated in a continuous, narrow band at the junctions, and electron microscopy revealed normal tongue-in-groove (TIG) structures (cellular process bordered on three sides by one from another cell) and TJs that are indistinguishable between normal donor and SCLS HDMECs (quantification, Fig. 2, C, E, and F). Additional staining revealed similar patterns of basal distribution for other junctional proteins, including ZO-1, CD144 (VE-cadherin), CD31 (PECAM-1), and JAM-A as well as staining for cortical actin fibers (Fig. S1). JAM-C staining was not discernable in HDMEC cultures. The response of the SCLS HDMECs to G-protein–coupled receptor agonists such as histamine and thrombin were also indistinguishable from normal ECs, with rapid declines and rapid recovery of transendothelial electrical resistance (TEER), a continuous, real-time, noninvasive measure of paracellular leak (Fig. 3 A). After 6 h of exposure to low-dose (0.5 ng/ml) TNF, both SCLS and normal donor HDMECs showed a similar pattern of disruption of staining for ZO-1, CD144, and CD31, and cortical actin showed disruption of cortical actin staining, although changes in JAM-A were not apparent (Fig. 2, B–F; and Fig. S1). By 24 h, significant recovery of contiguous membrane claudin-5 staining, TJs, and TIG features was observed in the normal donor HDMECs was noticeably absent in SCLS HDMECs. Other junctional markers showed less evidence of recovery compared with basal staining in both cell types (Fig. S1). When assessed by electrical cell-substrate impedance sensing (ECIS), SCLS HDMECs demonstrate both an accelerated rate of decrease in TEER in response to TNF stimulation (Fig. 3 B) and a profoundly compromised ability to recover barrier function after treatment with TNF compared with normal donor HDMECs. (Fig. 3 C). Similar differences were observed between SCLS HDMECs and normal donor HDMECs when cultures were subjected to treatment with IL-1β or Escherichia coli–derived LPS (Fig. S2).
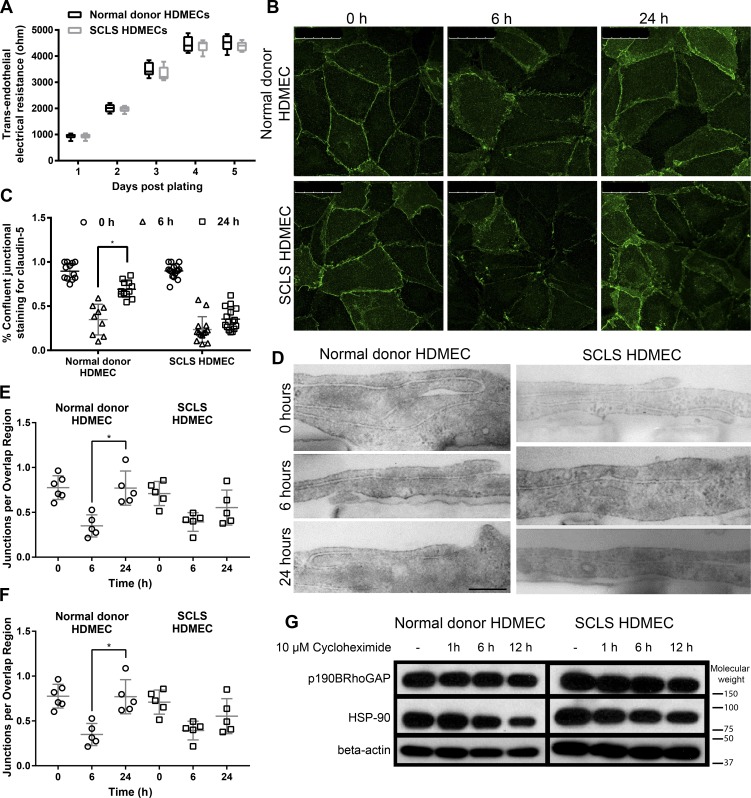
Figure 2.
Structural and functional comparison of HDMECs from the SCLS patient and a normal donor. (A) Acquisition of TEER with days in the culture for SCLS HDMECs compared with normal donor HDMECs. Data were compiled from three experiments. (B) Confocal microscopy of claudin-5. Postconfluent HDMEC monolayers organize claudin-5 into a thin peripheral band along cell junctions coincident with TJ formation (left). Treatment with 0.5 ng/ml TNF disrupts the claudin-5 pattern similarly between normal donor and SCLS HDMECs at 6 h (middle), and the recovery of junctional staining is more pronounced in the normal donor HDMECs after 24 h (right). Data are representative of three experiments. Bars, 50 µm. (C) Quantification of confluent claudin-5 junctional staining indicates significantly decreased recovery in the SCLS HDMECs. *, P < 0.05. (D) Electron microscopy of cellular junctions. Normal donor and SCLS patient HDMECs demonstrate similar patterns of cellular overlap (TIG) and numbers of TJs before stimulation with 0.5 ng/ml TNF, after 6 h both cell types display similar disruptions, and after 24 h the normal donor cells have recovered more overlap and TJs. Bars, 500 nm. (E and F) Quantification of electron microscopy reveals significantly decreased overlap in the SCLS cells (E) and significant recovery of TJ in the normal donor but not the SCLS cells (F). *, P < 0.05. Data are representative of two experimental replicates. (G) Stability of p190BRhoGAP. Normal donor and SCLS HDMECs were treated with 10 μM cycloheximide to maintain high levels of p190BRhoGAP. Data are expressed as means ± SDs.
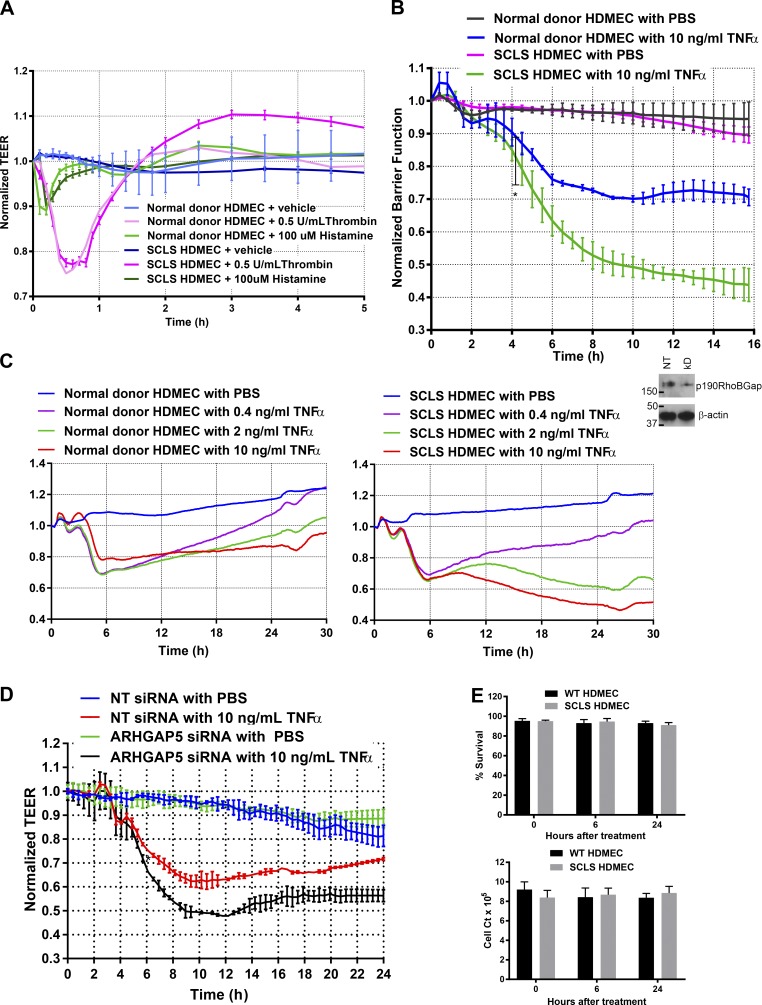
Figure 3.
Comparison of normal donor and SCLS HDMECs barrier function by using TEER. (A) Normal donor HDMECs and SCLS HDMECs demonstrate similar ECIS responses to stimulation with the G-protein–stimulating molecules histamine and thrombin. Data are from three experiments. There were no significant differences between normal donor and SCLS HDMECs. (B) There was a dramatic decrease in TEER (indicting increased permeability) in the SCLS HDMECs compared with normal donor HDMECs in response to stimulation with 10 ng/ml TNF. Significant differences between normal donor and SCLS HDMECs are noted at 4.5 h (*, P < 0.05). Data are from a collection of four separate experiments. (C) SCLS HDMECs demonstrate a prolonged barrier decrease in a dose-response relationship to TNF. Stimulation with all doses of TNF revealed increased permeability and delayed barrier recovery. Data are from two separate experiments (D) An ∼50% siRNA knockdown of p190BRhoGAP, assessed by immunoblotting, in normal donor HDMECs suggests p190BRhoGAP produces a similar dysfunction defect in barrier recovery as seen in the SCLS HDMECs by ECIS. Knockdown of the protein product of ARHGAP5, p190BRhoGAP by ∼50% with siRNA. Significant differences between nontargeting and ARHGAP5 siRNA-treated HDMECs are noted at 5 h. Data are from a collection from three experiments. (E) Viability and cell counts. Differences in TEER are not due to cell death (left) or cell number (right). There were no significant differences between cell types. Compiled data are from two experiments. Data are expressed as means ± SDs.
Identification and confirmation of a causative single protein mutation
The experiments with the SCLS HDMECs implied that the cause of systemic capillary leak in this pediatric patient was EC-intrinsic and related to an altered response to TNF and similarly signaling mediators. To identify a possible basis for this phenotype, we performed whole-exome sequencing on both the patient and the patient’s mother (the father declined to participate). Single-nucleotide variations (SNVs) were filtered for novelty in the Exome Aggregation Consortium (ExAC) database and absence in the healthy mother. In addition, we restricted our attention to genes known to be expressed in HDMECs by reference with an RNA whole-transcriptome dataset (not depicted). The 12 most likely disease-causing gene targets identified were ACAN, AGAP11, ARHGAP5, BCR, COMMD4, KSR1, PCDHGA12, RGPD5, STAM, TTN, and TYRO3. Each candidate gene was screened by using siRNA knockdown and for effects on TNF-mediated alterations in TEER. Only an siRNA knockdown of the protein p190BRhoGAP, encoded by the gene ARHGAP5, produced the same ECIS phenotype as observed in the SLCS HDMECs (Fig. 3 D). The patient, but not his mother, had an SNV, adenosine 2,285 to thymidine, in one copy of the ARHGAP5 gene on chromosome 14 (position 32,562,160 of genome build 37), as confirmed by Sanger sequencing. This SNV resulted in a single amino acid change, changing aspartic acid 762 to valine. This residue change is predicted to be very deleterious to protein function by PolyPhen2 (0.55, P), SIFT (0.04, D), LRT (0, D), and MutationTaster (1, D) scores and occurs at a site that is highly conserved among humans and other species. Unfortunately, the protein structure of p190BRhoGAP is not known, and the protein lacks sufficient similarity to proteins of known structure to reliably model the residue’s specific functional impact. The mutation may render a functional haploinsufficiency and/or act as a dominant negative mutation. Stability of p190BRhoGAP does not appear to be affected by this mutation (Fig. 2 G). A CRISPR/Cas9 approach to introduce or rescue a specific mutation in HDMECs is not possible because HDMECs lack the replicative capacity to allow for single-cell cloning. It is worth noting that, although several studies have been published on the function of p190ARhoGAP (also confusingly referred to as p190RhoGAP) in regulating endothelial permeability (Li et al., 1997; Su et al., 2003), this protein is encoded by the gene ARHGAP35 and shares only ∼50% sequence identity with the protein p190BRhoGAP identified as altered in our patient. The gene for ARHGAP35 was not mutated in our patient.
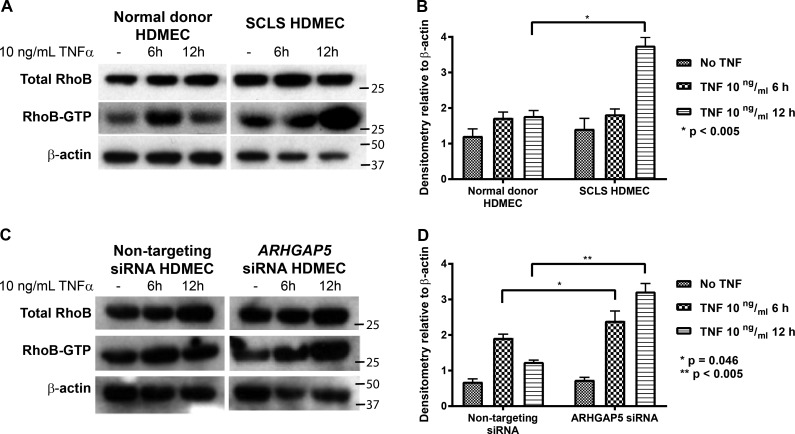
The exact functions of p190BRhoGAP are unknown, although it does not associate with RhoA and may associate with Rnd proteins (Wennerberg et al., 2003). Generally, Rho GTPase activating proteins (RhoGAPs) stimulate GTP hydrolysis by and inactivation of one or more of the three Ras homologue gene family members expressed in humans (RhoA, RhoB, and RhoC). RhoA has been implicated in alterations of barrier function by thrombin, but neither its abundance nor its activation is altered by TNF in HDMECs (not depicted). In contrast, both the abundance of RhoB transcript (Table 1) and activation of RhoB protein are increased by TNF treatment of HDMECs. RhoB has recently been shown to have a unique C-terminal region and may have specialized functions not shared by RhoA or RhoC (Vega and Ridley, 2016). Importantly, activated RhoB has been specifically implicated as an inhibitor of EC barrier recovery by sequestering Rac1 away from intercellular junctions in human umbilical vein ECs stimulated by thrombin (Marcos-Ramiro et al., 2016). Consistent with this study, we found increased active RhoB and prolonged activation of RhoB in response to TNF in SCLS HDMECs compared with control HDMECs (Fig. 4, A and B). Interestingly, the net increase of RhoB transcript determined by RNA sequencing (Table 1) and total RhoB protein in normal donor HDMECs before and after TNF treatment is less pronounced than the increase in activated (GTP-bound) RhoB (Fig. 4 A). The absolute levels of RhoB protein and active RhoB varied among different control HDMEC cultures at baseline, but all showed qualitatively similar responses to TNF, with an early increase at 6 h that was largely unchanged or declining by 12 h. Only SCLS HDMECs showed significantly more active RhoB at 12 h than 6 h of TNF treatment. The sustained or increasing activation of RhoB correlates with the defective recovery of barrier function observed on ECIS. A similar pattern of prolonged RhoB activation was observed after the siRNA knockdown of p190BRhoGAP in normal donor HDMEC (Fig. 4, C and D). The rescue of SCLS HDMECs by overexpression of wild-type protein would provide further support for the causal role of our patient’s mutation of ARHGAP5 both as a specific RhoB GAP and as the etiological basis of the capillary leak, but the limited supply of these cells and the availability of suitable cDNA constructs precluded such an analysis.
Table 1. Whole-transcriptome analysis of Rho proteins in normal HDMECs before and after stimulation with 10 ng/ml TNF for 6 h.
| RHO | Number of reads | Log2 fold change | P-value | |
|---|---|---|---|---|
| Vehicle | 10 ng/ml TNF for 6 h | |||
| RHOA | 319 | 335 | 0.072 | 0.43 |
| RHOB | 560 | 1,729 | 1.62 | <0.05 |
| RHOC | 248 | 368 | 0.574 | <0.05 |
Figure 4.
RhoB activation in isolated and siRNA knockdown ECs. (A) Representative set of Western Blot results from RhoB-GTP rhotekin pulldown assays of SCLS and normal donor HDMECs stimulated with TNF. Total RhoB and β-actin was obtained from replicated wells. (B) Quantification of four experiments comparing RhoB-GTP with β-actin in SCLS and normal donor HDMECs. (C) Representative set of Western Blot results from RhoB-GTP rhotekin pulldown assays of normal donor HDMECs treated with nontargeting or ARHGAP5-targeted siRNA stimulated with TNF. Total RhoB and β-actin was obtained from replicated wells. (D) Quantification of four experiments comparing RhoB-GTP to β-actin in SCLS and normal donor HDMECs treated with nontargeting or ARHGAP5 siRNA. Data are expressed as means ± SDs.
In conclusion, we report a case of a pediatric patient with fatal SCLS associated with a mutation in p190BRhoGAP and show that this protein functions to terminate capillary leak in response to TNF through inactivation of signaling by RhoB in HDMECs. Although TNF-mediated permeability was evaluated in this study, similar responses of SCLS HDMECs to IL-1β and LPS were observed, each of which caused HDMECs to disrupt TJs in a process involving NF-κB-dependent new gene expression, including RhoB (Clark et al., 2015). The defect discovered in this patient may have revealed a potential target for controlling inflammation-associated capillary leak induced by severe illness in the general population.
Materials and methods
Autopsy
The autopsy was conducted at an interval of 19 h post-mortem and began with the standard, complete external examination. A comprehensive internal assessment followed, which included both in situ and ex situ reviews of all organs in fresh and formalin-fixed states, respectively. The autopsy concluded with the histological evaluation of tissues representing all major organs. Immunostains and special stains were performed on an as-needed basis. ECs were identified by anti–CD31 antibody (Agilent) shown in Fig. 1 that was used via the standard protocol used for all surgical pathology specimens at our institution.
Isolation of HDMECs
All human cells were isolated under protocols approved by the Yale Institutional Review Board. Control HDMECs were isolated from normal human skin collected from reduction mammoplasties or abdominoplasties obtained as discarded and deidentified tissue from Yale-New Haven Hospital, New Haven, CT. Patient-derived SCLS HDMECs were cultured by the same approach from abdominal skin obtained at necropsy. Fresh skin was cut into 3 × 10–cm sections, stretched flat, and sectioned horizontally by using a dermatome with a 0.016-in-depth gauge and guard (Weck). After a 30-min incubation in dispase (Collaborative Biomedical Products) in a 37°C water bath, the epidermis was peeled off, and cells from both sides of the underlying dermis were gently scraped into RPMI 1640 media (Life Technologies) and filtered through a 70-pM nylon mesh. The filtrate, containing single cells, was washed once in EGM2-MV (Lonza) and plated onto tissue culture plastic (Becton Dickinson) precoated with human plasma fibronectin (EMD Millipore) and cultured at 37°C in a humidified 5% CO atmosphere. When the cultures reached ∼50% confluence, the cells were suspended with trypsin and positively selected for CD31 with magnetic beads (Miltenyi) per the manufacturer’s protocol. The selected cells were cultured and serially passaged in 0.1% gelatin-coated (Sigma) tissue culture flasks. By FACS analysis, the cells appeared uniformly positive for CD31, CD34, and endoglin. Cultured HDMECs express both the lymphatic EC markers Prox-1 and podoplanin and will express blood vascular EC markers E-selectin and VCAM-1 on treatment with TNF. Aliquots of HDMECs are cryopreserved in 10% DMSO (Sigma) in FBS (VWR) at the third passage and may be serially passaged for an additional 5 passages. All experiments in this manuscript comparing normal donor HDMECs to SCLS HDMECs used cells at identical passage levels.
Whole-exome sequencing
DNA was prepared from venous blood samples of our SCLS patient and his mother under Yale’s Institutional Review Board protocol with appropriate informed assent and consent. Exome sequencing was performed by capture on the NimbleGen 2.1 Exome reagent followed by 74-base-paired end sequencing on an Illumina platform (HiSeq 2000) to high coverage (each targeted base was read by a mean of >80 independent reads in each subject) as previously described (Romberg et al., 2014). Sequences were aligned to NCBI Build 36 of the human genome and SNVs and indel calls were assigned quality scores by using SAMtools and annotated for novelty (by using ExAC, Yale, 1000 genomes, and NHLBI exome databases), for impact on encoded proteins, and for conservation of variant position, all as previously described (Romberg et al., 2014). A total of 76,139 SNVs were identified in the patient and analyzed. SNV analysis was conducted in stepwise fashion: (1) Limit to exonic SNVs; (2) Delete silent SNVs; (3) Filter by novelty in the ExAC genome database; (4) Analyze all SNVs that result in loss of protein function; (5) Elimination of SNVs found in the patient’s asymptomatic mother; and (6) Compare with RNA sequencing from normal donor HDMEC under similar culture conditions and before and after TNF exposure. Sanger sequencing was performed to confirm the mutation of the presence of the ARHGAP5 SNV in the patient and absence in the patient’s mother.
TEER measurements
TEER of HDMEC monolayers was assessed by ECIS (8- or 96-well arrays, catalog numbers 96W20idf or 8W10E+, respectively, both with polyethylene terephthalate; from Applied BioPhysics) stimulated with TNF (Life Technologies), IL-1β (PeproTech), and LPS (Sigma) as described (Kluger et al., 2013). Figures illustrate normalized TEER values (where the value of 1.0 represents the basal TEER measurement immediately before adding cytokine). Data were recorded by an ECIS Z-theta instrument controlled by a Dell personal computer equipped with ECIS software (Applied BioPhysics).
siRNA inhibition of gene expression
HDMECs plated on gelatin-coated (Sigma) 6-well plates (Flacon, Corning) at 50% confluence were transfected with Lipofectamine RNAiMAX (Invitrogen) complexed to either of two different siRNA sequences that specifically targeted to ARHGAP5 per the manufacturers protocol. The siRNA sequences used were commercially available (Dharmacon D-009580-17 and D-009580-18 for ARHGAP5 and D-001210-02-05 for nontargeting control). Both targeting siRNAs gave qualitatively similar results in preliminary experiments (Fig. S2), and the more effective of the two, D-009580-17, was used in all subsequent experiments. In brief, siRNA complexes of Lipofectamine (Invitrogen) at 50 µg/ml and siRNA at 100 nM were prepared in Opti-MEM I Reduced Serum Medium (Invitrogen) and then diluted fivefold in Opti-MEM to yield a final siRNA concentration of 20 nM. The siRNA-Lipofectamine was then added to HDMECs cultures and incubated for 2 h at 37°C. Fresh medium (EGM-2MV, Lonza) was then added overnight, and cells were retransfected 24 h later. After resting for 24 h, HDMECs were then trypsinized and seeded into gelatin-coated ECIS 8-chamber arrays (Applied BioPhysics, catalog #8W10E+) at 100,000 cells per well or maintained in the same culture ware for protein analysis.
Immunoblot analysis of protein expression and rhotekin pull-down assays
For immunoblot analyses, HDMECs cultured as described in C12 or C24 plastic wells (Falcon, Corning) were prepared for electrophoresis with SDS-PAGE gels (Bio-Rad), transferred onto polyvinylidene fluoride filters (Millipore), and immunoblotted as previously described (Kluger et al., 2013). Antibodies for RhoB (Cell Signaling, Cell Biolabs), β-actin (ThermoFisher), HSP90 (ThermoFisher), and p190BRhoGAP (Bethyl) were used. Immunoblot images display bands of interest and controls from the same contiguous immunoblots with background levels minimized by adjusting all pixels equally with ImageJ 1.50i software (National Institutes of Health). For immunoblotting for p190BRhoGAP stability, confluent HDMEC monolayers were treated with 10 μM cycloheximide (Sigma) and washed with phosphate buffered saline (Lonza) twice before collection with SDS-PAGE–reducing Lameli buffer (Bio-Rad).
Rhotekin pull-down assays were performed by using RhoB Activation Assay kits (Cell Biolabs). HDMECs cultured in C12 or C24 plastic wells (Flacon, Corning) were lysed with the buffer contained in the activation assay kit and either flash frozen in liquid nitrogen or processed immediately. Incubation with rhotekin-agarose beads was performed with agitation for 1 h at 4C. The beads were pelleted by centrifugation and washed with the kit buffer three times. Samples were either frozen at −80°C or processed immediately by mixing with 30 μL of standard SDS-PAGE–reducing Laemmli buffer (Bio-Rad) and placed in a boiling water bath for 10 min and run on a protein gel as described above for immunoblotting.
Immunofluorescence microscopy and morphometric measurements
Treated HDMEC monolayers grown on gelatin-coated glass were washed briefly and fixed in 95% ethanol for 30 min at 4°C. For two-color immunofluorescence imaging, monolayers were incubated overnight in anti–caludin-5 antibody (Invitrogen), ZO-1 (ThermoFisher), CD144 (eBiosciences), CD31 (Life Technologies), JAM-A (R&D Biosystems), and JAM-C (R&D Biosystems) and diluted in Tris-buffered solution/0.2% Triton-X-100/5% normal donkey serum. Donkey Alexa488 and Alexa594 Donkey secondary antibodies along with phalloidin-488 (Life Technologies) were used to detect primary antibody and β-actin. Glass coverslips were mounted for immunofluorescence analysis in ProLong mounting media (Invitrogen). Randomly selected (minimum of five images per experimental condition) fluorescence photomicrographs were collected by using a Zeiss Axiovert fluorescence microscope and Plan-APOCHROMAT, 63× oil objective (Zeiss) with an ORCA-ER digital camera (Hamamatsu Photonics).
Electron microscopy
ECs, isolated and propagated as described above in the Isolation of HDMECs section, were seeded onto fibronectin-coated high-density 0.4-mm-pore-size, six-well-format cell culture inserts (BD Biosciences) and prepared for electron microscopy as previously described (Kluger et al., 2013). TJ ultrastructures were identified by transmission electron microscopy (JEOL) imaging as sites where membranes converge as electron-dense material indicative of intracellular cytoplasmic plaque proteins. Juxtapositions of plasma membrane processes from neighboring ECs were scored as TIG structures where at least one layer has a visible blunt end surrounded on three sides by membrane protrusions originating from an adjacent cell.
Statistics
Data are expressed as means ± SDs. Statistical analyses were performed by using GraphPad Prism software (V7.0b, GraphPad). Unpaired t test or Mann-Whitney U tests were used to make statistical comparisons between normal donor and SCLS HDMECs in different treatment groups. In all experiments, P < 0.05 was considered statistically significant.
Experimental guidelines approval statement
All experiments in this work were approved by Yale University’s Human Investigation Committee. Additional assent and consent for whole-exome sequencing studies were obtained and stored by the Pediatric Genomic Discovery Center as dictated by Yale University. No animals were used in this work.
Online supplemental material
Fig. S1 shows confocal staining for ZO-1, CD144, JAM-A, CD31, and β-actin of patient and normal donor HDMECs treated with 0.5 ng/ml TNF. Fig. S2 shows the TEER response of normal donor and patient HDMECs treated with 100 ng/ml LPS and 1 ng/ml IL-1 as well as confirmatory siRNA knockdown of p190BRhoGAP in normal donor HDMECs.
Supplementary Material
Acknowledgments
The authors thank the patient and family, who are the inspiration for this study. We thank the Yale Center for Genome Analysis (YCGA) for exome sequencing support including the assistance by James Knight for sequence alignment algorithms.
This work is supported by the National Institutes of Health grants R01HL36003 to J.S. Pober and M.S. Kluger, R01HD081379 and R33HL120783 to M.K. Khokha, and T32HD068201 to R.W. Pierce. M.K. Khokha is a Mallinckrodt Scholar. The Pediatric Genomics Development Program is supported by Sara and Jeffery R. Buell. We thank the Yale Pathology Department and the medical care team, especially Drs. Tanya Murtha and Raffaella Morotti, for facilitating data and sample collection.
The authors declare no competing financial interests.
Author contributions: R.W. Pierce cared for the patient before his demise, coordinated tissue collection, and conducted research studies. R.W. Pierce and J.S. Pober designed the research studies, analyzed the data, and wrote the manuscript. J.P. Lavik and A. Huttner conducted the pathological and electron microscopy studies. J. Merola and M.S. Kluger conducted experiments and helped analyze the data. M.K. Khokha conducted whole-exome sequencing and analyzed these data.
Note Added in Proof
Since acceptance of our manuscript, possible functional significance has been assigned to the SNV reported in this manuscript. As reported by Stiegler and Boggon (2017. Nat. Commun. https://doi.org/10.1038/s41467-017-00483-x), the D762V mutation identified in this patient occurs at the C-terminal end of the first of two newly identified pseudoGTPase domains within p190BRhoGAP.
Footnotes
Abbreviations used:
- EC
- endothelial cell
- ECIS
- electrical cell-substrate impedance sensing
- ExAC
- Exome Aggregation Consortium
- HDMEC
- human dermal microvascular EC
- SCLS
- systemic capillary leak syndrome
- SNV
- single-nucleotide variation
- TEER
- transendothelial electrical resistance
- TIG
- tongue-in-groove
- TJ
- tight junction
References
- Assaly R., Olson D., Hammersley J., Fan P.S., Liu J., Shapiro J.I., and Kahaleh M.B.. 2001. Initial evidence of endothelial cell apoptosis as a mechanism of systemic capillary leak syndrome. Chest. 120:1301–1308. 10.1378/chest.120.4.1301 [DOI] [PubMed] [Google Scholar]
- Clark P.R., Kim R.K., Pober J.S., and Kluger M.S.. 2015. Tumor necrosis factor disrupts claudin-5 endothelial tight junction barriers in two distinct NF-κB-dependent phases. PLoS One. 10:e0120075 10.1371/journal.pone.0120075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson B., Thompson D., Horwith M., and Luckey E.H.. 1960. Cyclical edema and shock due to increased capillary permeability. Am. J. Med. 29:193–216. 10.1016/0002-9343(60)90018-8 [DOI] [PubMed] [Google Scholar]
- Hsu P., Xie Z., Frith K., Wong M., Kakakios A., Stone K.D., and Druey K.M.. 2015. Idiopathic systemic capillary leak syndrome in children. Pediatrics. 135:e730–e735. 10.1542/peds.2014-2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita Y., Kasaoka S., Fujita M., Oshima C., Kawamura Y., Tsuruta R., and Maekawa T.. 2010. Synchronized changes in serum vascular endothelial growth factor during the clinical course of chronic systemic capillary leak syndrome. Intern. Med. 49:791–794. 10.2169/internalmedicine.49.2929 [DOI] [PubMed] [Google Scholar]
- Kluger M.S., Clark P.R., Tellides G., Gerke V., and Pober J.S.. 2013. Claudin-5 controls intercellular barriers of human dermal microvascular but not human umbilical vein endothelial cells. Arterioscler. Thromb. Vasc. Biol. 33:489–500. 10.1161/ATVBAHA.112.300893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zhang B., and Zheng Y.. 1997. Structural determinants required for the interaction between Rho GTPase and the GTPase-activating domain of p190. J. Biol. Chem. 272:32830–32835. 10.1074/jbc.272.52.32830 [DOI] [PubMed] [Google Scholar]
- Marcos-Ramiro B., García-Weber D., Barroso S., Feito J., Ortega M.C., Cernuda-Morollón E., Reglero-Real N., Fernández-Martín L., Durán M.C., Alonso M.A., et al. . 2016. RhoB controls endothelial barrier recovery by inhibiting Rac1 trafficking to the cell border. J. Cell Biol. 213:385–402. 10.1083/jcb.201504038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg N., Al Moussawi K., Nelson-Williams C., Stiegler A.L., Loring E., Choi M., Overton J., Meffre E., Khokha M.K., Huttner A.J., et al. . 2014. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat. Genet. 46:1135–1139. 10.1038/ng.3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Agati J.M., and Parsons S.J.. 2003. p190RhoGAP is cell cycle regulated and affects cytokinesis. J. Cell Biol. 163:571–582. 10.1083/jcb.200308007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega F.M., and Ridley A.J.. 2016. The RhoB small GTPase in physiology and disease. Small GTPases.:1–10. 10.1080/21541248.2016.1253528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K., Forget M.-A., Ellerbroek S.M., Arthur W.T., Burridge K., Settleman J., Der C.J., and Hansen S.H.. 2003. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr. Biol. 13:1106–1115. 10.1016/S0960-9822(03)00418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Ghosh C.C., Patel R., Iwaki S., Gaskins D., Nelson C., Jones N., Greipp P.R., Parikh S.M., and Druey K.M.. 2012. Vascular endothelial hyperpermeability induces the clinical symptoms of Clarkson disease (the systemic capillary leak syndrome). Blood. 119:4321–4332. 10.1182/blood-2011-08-375816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Nagarajan V., Sturdevant D.E., Iwaki S., Chan E., Wisch L., Young M., Nelson C.M., Porcella S.F., and Druey K.M.. 2013. Genome-wide SNP analysis of the Systemic Capillary Leak Syndrome (Clarkson disease). Rare Dis. 1:e27445 10.4161/rdis.27445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Ghosh C.C., Parikh S.M., and Druey K.M.. 2014. Mechanistic Classification of the Systemic Capillary Leak Syndrome: Clarkson Disease. Am. J. Respir. Crit. Care Med. 189:1145–1147. 10.1164/rccm.201310-1746LE [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.