The molecular mechanisms that regulate AID mutator activity at off-target genes are not well characterized. Mu et al. show AID phosphorylation dynamically controls activity at Myc and other sites. Pharmacological induction of AID phosphorylation leads to increased mutations, double strand breakss and translocations.
Abstract
Activation-induced cytidine deaminase (AID) is a mutator enzyme that targets immunoglobulin (Ig) genes to initiate antibody somatic hypermutation (SHM) and class switch recombination (CSR). Off-target AID association also occurs, which causes oncogenic mutations and chromosome rearrangements. However, AID occupancy does not directly correlate with DNA damage, suggesting that factors beyond AID association contribute to mutation targeting. CSR and SHM are regulated by phosphorylation on AID serine38 (pS38), but the role of pS38 in off-target activity has not been evaluated. We determined that lithium, a clinically used therapeutic, induced high AID pS38 levels. Using lithium and an AID-S38 phospho mutant, we compared the role of pS38 in AID activity at the Ig switch region and off-target Myc gene. We found that deficient pS38 abated AID chromatin association and CSR but not mutation at Myc. Enhanced pS38 elevated Myc translocation and mutation frequency but not CSR or Ig switch region mutation. Thus, AID activity can be differentially targeted by phosphorylation to induce oncogenic lesions.
Introduction
Activation-induced cytidine deaminase (AID) initiates antibody diversification by introducing U:G mismatches in the Ig genes of activated B lymphocytes. During somatic hypermutation (SHM), AID targets the variable regions of both the Ig heavy and light chain genes, inducing high point mutation rates (10−2 to 10−3 mutations/bp per generation; Di Noia and Neuberger, 2007). In class switch recombination (CSR), AID targets the noncoding switch regions upstream of constant region exons, inducing double-strand break (DSB) substrates for CSR. Subsequent error-prone processing of uracil by uracil-DNA glycosylase (UNG) base excision repair pathway or, alternatively, mismatch repair MSH2/6 pathways complete SHM and CSR (Stavnezer et al., 2008; Methot and Di Noia, 2017).
The mechanisms that target AID are not Ig exclusive, and AID has widespread off-target association with actively transcribed regions throughout the genome (Chandra et al., 2015; Casellas et al., 2016). AID-dependent mutagenesis and chromosome rearrangement occur at off-target loci, which include oncogenes that are recurrent translocation partners in B cell tumors (Alt et al., 2013; Robbiani and Nussenzweig, 2013). One such example, the oncogenic Myc/Igh translocation, is potentiated by AID-initiated DSB at both Myc and Igh genes (Ramiro et al., 2004; Robbiani et al., 2008). However, even with promiscuous AID association, off-target mutation rates are low compared with Ig genes, demonstrating a differential between occupancy and mutation (Liu et al., 2008). AID chromatin binding does not directly correspond to DNA damage (Yamane et al., 2011; Hakim et al., 2012; Matthews et al., 2014), suggesting that factors beyond AID recruitment influence the targeting of AID mutator activity.
AID activity is modulated in vivo by phosphorylation on several residues including serine 3, serine 38, and threonine 140 (McBride et al., 2008; Gazumyan et al., 2011; Le and Maizels, 2015). The best-characterized modification is phosphorylation on AID serine 38 (pS38; Basu et al., 2005). Only a small percentage of total cellular AID is pS38 modified, but it is enriched on chromatin-associated AID (McBride et al., 2006). The role of pS38 in CSR and SHM has been defined with AID serine 38 mutated to alanine (AIDS38A) knock-in mice. B cells from these mice support CSR and SHM at 20% of the level of WT mice, demonstrating that pS38 regulates AID activity at the Ig variable and switch regions (McBride et al., 2008; Cheng et al., 2009). However, off-target AID activity has not been evaluated in these mice.
S38 is a consensus site for several kinases (McBride et al., 2008), including the cAMP-dependent protein kinase, PKA (Basu et al., 2005). PKA associates through its regulatory subunit at the Ig switch regions, potentiating AID phosphorylation and activity there. Consistent with this, PKA inhibitors or hypomorphic PKA mutants impair CSR (Pasqualucci et al., 2006; Vuong et al., 2009). DNA damage and activation of ataxia telangiectasia mutated (ATM) facilitate a pS38 positive feedback loop to facilitate CSR (Vuong et al., 2013). Although the switch regions provide an environment rich in DSBs and ATM activation, a role for ATM and PKA in regulating pS38 at Ig variable regions or off-target genes is not clear.
With an incomplete understanding of signaling pathways that affect AID phosphorylation, we sought to identify pharmacological inhibitors that alter AID pS38. In our studies, we discovered that lithium salts significantly increase pS38. Lithium is used clinically to treat bipolar disorder, depression, and associated mental health disorders (Goodwin et al., 2007), but also modulates several inflammatory and immune functions (Chiu et al., 2013; Maddu and Raghavendra, 2015). Lithium is an inhibitor of glycogen synthase kinase 3 (GSK3; Ryves and Harwood, 2001), which was our original rationale for analysis. However, lithium modulates other signaling pathways and transcription factors unrelated to GSK3 (Lenox and Wang, 2003; Chiu et al., 2013). In this study, we use lithium as a tool to investigate the consequence of altered AID pS38 on AID activity. Together with analysis of AIDS38A B cells, we defined the role of pS38 in mutation activity at the Ig switch region, the Myc oncogene, and genome-wide off-targets.
Results and discussion
Lithium treatment induces AID serine 38 phosphorylation
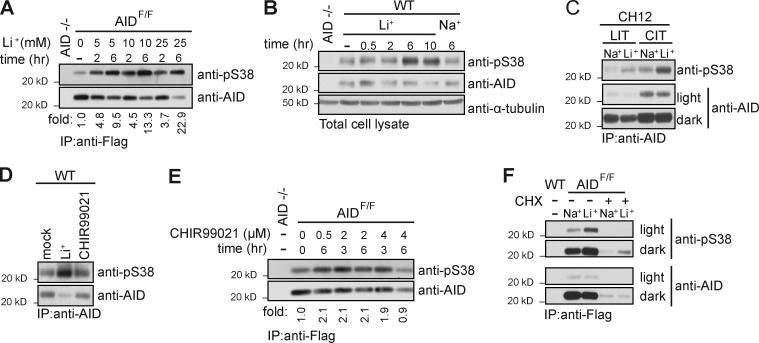
AID is phosphorylated on S38, and this modification regulates AID activity in SHM, CSR, and gene conversion (Chatterji et al., 2007; McBride et al., 2008; Cheng et al., 2009). Anti-pS38 antibodies have been produced and specifically detect pS38 on AID (Fig. S1 A; McBride et al., 2006, 2008). To determine the impact of lithium on AID pS38 levels, we examined AID from splenocytes stimulated to undergo CSR. We initially used B cells from Flag-tag knock-in AID (AIDF/F) mice that express functional AID at physiological levels (Pavri et al., 2010). B cells were activated with LPS and IL-4 for 3 d before lithium acetate (LiAc) treatment. We found that LiAc increased pS38 in a time- and dose-dependent manner (Fig. 1 A). This effect was lithium specific, as both LiCl and LiAc (Fig. S1 B) increased pS38 levels, whereas NaCl, used as a control, did not (Fig. 1 B). LiCl induced pS38 in activated primary WT B cells (Fig. 1 B) and in the B cell line CH12 (Fig. 1 C). We concluded that lithium induces pS38 beyond physiological levels. Interestingly, the LiCl induced a small but consistent drop in total AID levels (Fig. S1 C).
Figure 1.
Lithium treatment induces AID Serine 38 phosphorylation. (A) Anti-pS38 and anti-AID immunoblot of Flag-AID purified with anti-Flag antibody from lysates of 3-d LPS- and IL-4–stimulated AIDF/F B cells treated with indicated concentration of lithium acetate (Li+) for indicated time before harvest. (B) Anti-pS38, anti-AID, and anti–α-tubulin immunoblot of cell lysates from WT B cells treated with 10 mM LiCl (Li+) or NaCl (Na+) as a control for the indicated time. (C) Anti-pS38 and anti-AID immunoblot (light and dark exposures) of AID immunoprecipitated from lysates of CH12 cells cultured with LPS (LIT) or CD40 (CIT) together with IL-4, and TGF-β and treated with 10 mM LiCl or NaCl for 6 h. (D) Representative anti-pS38 and anti-AID immunoblot of AID purified with anti-AID antibody from lysates of WT B cell lysates cultured for 3 d in LPS and IL-4. Cells were treated with 2 µM CHIR99021 or 25 mM LiCl (Li+) for 6 h before harvest. (E) Anti-pS38 and anti-AID immunoblot of Flag-AID purified with anti-FLAG antibody from cultured AIDF/F B cell lysates. Concentration and time of CHIR99021 treatment are indicated. (F) Immunoblot of anti-pS38 and anti-AID (light and dark exposures) of Flag-AID purified with anti-Flag antibody from cell lysates. WT or AIDF/F cultured B cells were treated with 10 µg/ml cycloheximide (CHX) 1 h before addition of 10 mM LiCl for 6 h. All panels representative of n = 3 independent experiments except F, n = 2.
Lithium is an inhibitor of GSK3 (half-maximal inhibitory concentration [IC50] 2 mM; Ryves and Harwood, 2001), and this action is thought to contribute to its mood-stabilizing effects. (Klein and Melton, 1996; Chiu et al., 2013). To determine whether GSK3 influences pS38 levels, we treated B cells with the specific GSK3 inhibitor CHIR99021 (IC50 10 nM; Ring et al., 2003). Although treatment with LiCl led to a substantial increase in pS38, CHIR99021 treatment did not (Fig. 1 D). The effect of CHIR99021 on pS38 was a modest ∼2-fold increase with varied time and inhibitor concentrations (Fig. 1 E). We concluded that lithium is not regulating pS38 through the GSK3 pathway. Lithium has pleiotropic effects in B cells; 12-h LiCl treatment altered phospho-sites on a small subset of proteins regulated by different kinases (Fig. S1 D). Lithium also modulates expression of several genes. To determine whether new protein synthesis is required for LiCl-mediated pS38 induction, we treated B cells with the ribosome translation inhibitor cycloheximide (CHX). 7-h CHX treatment led to a substantial drop in AID levels. However, LiCl treatment resulted in an ∼8-fold increase in pS38 signal regardless of CHX treatment (Fig. 1 F). We concluded that de novo protein synthesis is not required for LiCl to induce pS38.
AID serine 38 phosphorylation and chromatin association
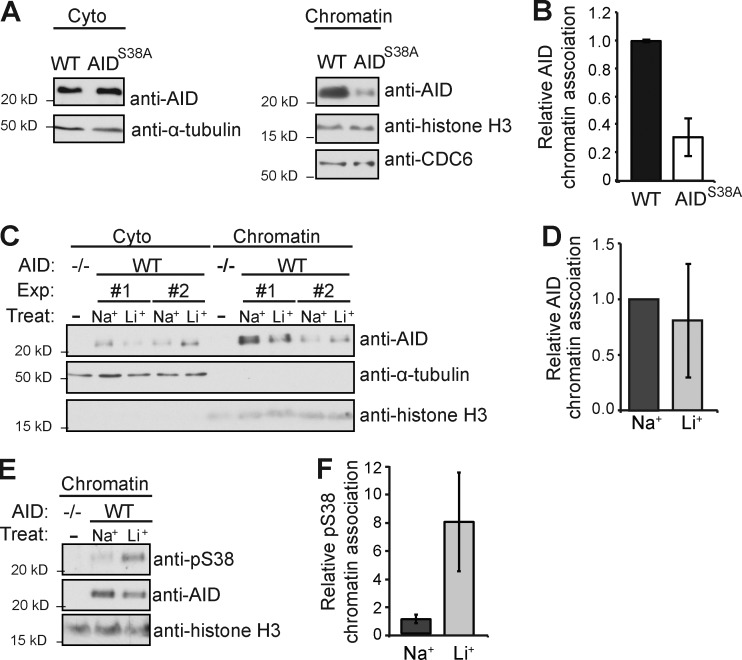
AID must shuttle to the nucleus and associate with chromatin to function. In B cells undergoing CSR, the majority of AID is cytoplasmic, and only a small percentage (5–10%) is associated with chromatin, which is enriched in pS38 (McBride et al., 2006). To determine whether pS38 influences AID chromatin association, we purified the chromatin fraction of B cells from WT or AIDS38A mice and determined the relative AID level (Fig. 2, A and B). We found that AIDS38A B cells displayed AID chromatin levels ∼30% those of WT cells, suggesting that pS38 promotes chromatin association of AID. To determine whether enhanced pS38 increased AID chromatin association, we analyzed cytosolic and chromatin fractions from LiCl-treated or control NaCl-treated B cells (Fig. 2, C and D). Although LiCl induced no significant change in AID chromatin level, LiCl did significantly increase the pS38 level of AID associated with chromatin (Fig. 2, E and F). These results suggest that a population of nonphosphorylated AID is normally associated with chromatin and that LiCl can enhance pS38 levels on chromatin.
Figure 2.
AID Serine 38 phosphorylation and chromatin association. (A) Representative anti-AID, anti–α-tubulin, anti–histone H3, and anti-CDC6 immunoblot of the cytoplasmic and chromatin fraction from WT or AIDS38A B cells cultured with LPS and IL-4 for 72 h. (B) Summary of relative chromatin levels of n = 5 independent experiments. Relative AID signal was normalized to histone H3 level, and WT was arbitrarily set to 1 for each experiment. (C) Representative anti-AID, anti–α-tubulin, and anti–histone H3 immunoblot of the cytosolic and chromatin fraction of LPS- and IL-4–stimulated B cells treated with 10 mM LiCl (Li+) or NaCl (Na+) as a control for 12 h. Two independent experiments (Exp) are shown. (D) Summary of relative AID chromatin level from n = 4 independent experiments. Relative AID signal was normalized to histone signal, and control (Na+) was set to an arbitrary value of 1 for each experiment. (B and D) Error bars are SE. (E) Representative anti-pS38, anti-AID, and anti–histone H3 immunoblot of the chromatin fraction from cultured AID−/− or WT B cells treated with 10 mM LiCl or NaCl for 12 h. (F) Summary of mean relative pS38 to AID signal from n = 3 independent experiments and SE are displayed.
Enhanced AID serine 38 phosphorylation does not increase CSR
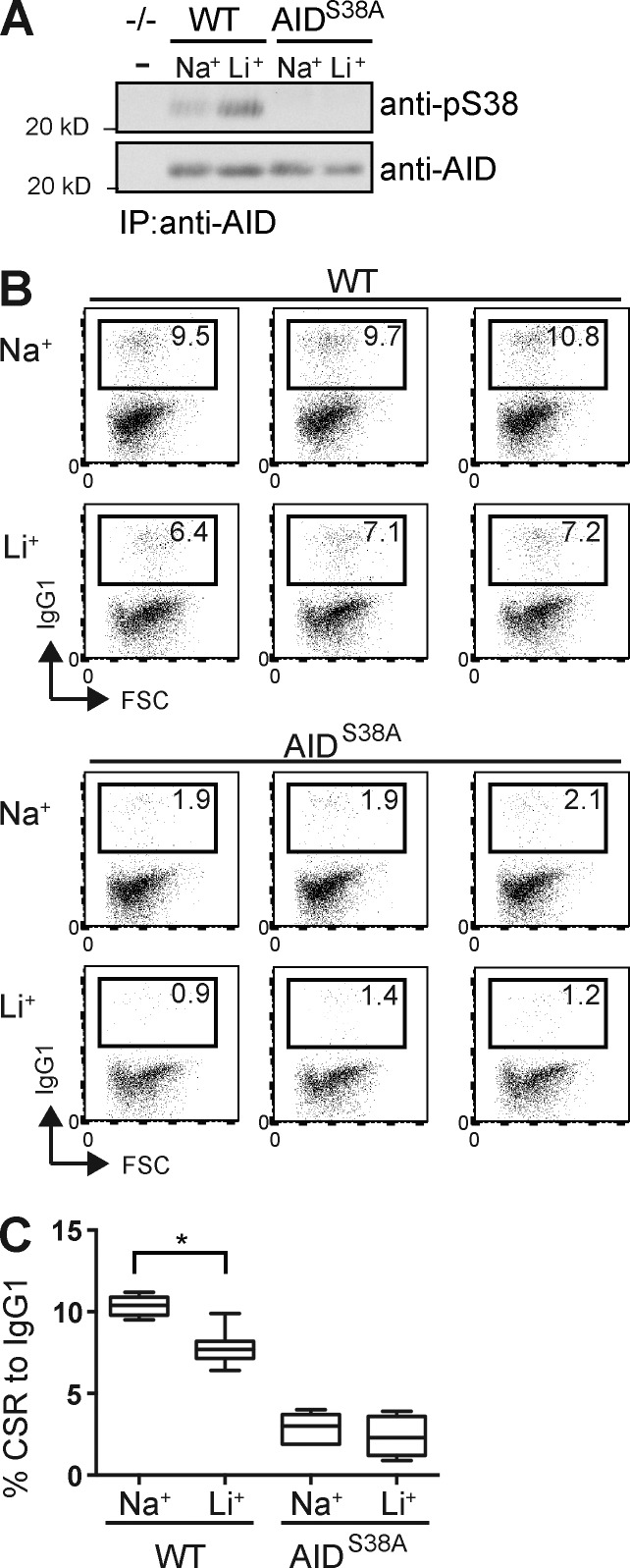
AID phosphorylation is required for full activity in CSR. B cells from AIDS38A B cells support CSR to IgG1 at ∼20% the rate of WT (McBride et al., 2008; Cheng et al., 2009). To determine whether higher pS38 increases CSR, we analyzed the effect of LiCl on CSR. To distinguish the effect of enhanced pS38 from pleotropic effects of LiCl, we compared CSR to IgG1 in B cells from WT and AIDS38A mice (Fig. 3 A). Naive B cells were activated with LPS and IL-4 for 3 d, with 10 mM LiCl or NaCl treatment for the final 12 h before analysis (Fig. 3 A). LiCl-treated WT cells displayed CSR to IgG1 at levels 75% those of controls (Fig. 3, B and C). As expected, AIDS38A B cells displayed CSR at rates 20–30% of WT (McBride et al., 2008; Cheng et al., 2009). The addition of LiCl treatment resulted in AIDS38A CSR at 80% of NaCl control (Fig. 3, B and C). We found that LiCl treatment of both WT and AIDS38A B cells resulted in similar CSR reduction compared with NaCl control. We concluded that increasing pS38 beyond physiological levels does not enhance CSR. The small, but significant, decrease in WT B cell CSR may be caused by pleiotropic effects because we observed a similar, although not significant, trend in AIDS38A B cells.
Figure 3.
Enhanced AID Serine 38 phosphorylation does not increase CSR. (A) Anti-pS38 and anti-AID immunoblot of AID purified with anti-AID antibody from lysates of B cells from WT, AIDS38A, or AID−/− mice. Cells were cultured with IL-4 and LPS for 72 h and treated with 10 mM LiCl (Li+) or NaCl (Na+) for 12 h before analysis. (B) Representative triplicate FACS plots of CSR to IgG1 in WT and AIDS38A B cells from A. Relative percentage of cells expressing IgG1 is in each plot. (C) Summary of CSR to IgG1 in activated B cells from n = 6 individual experiments each in triplicate. Error bars are SE. P-value (*, P < 0.001) was determined by a two-tailed t test assuming unequal variance.
AID serine 38 phosphorylation promotes Myc/Igh translocations
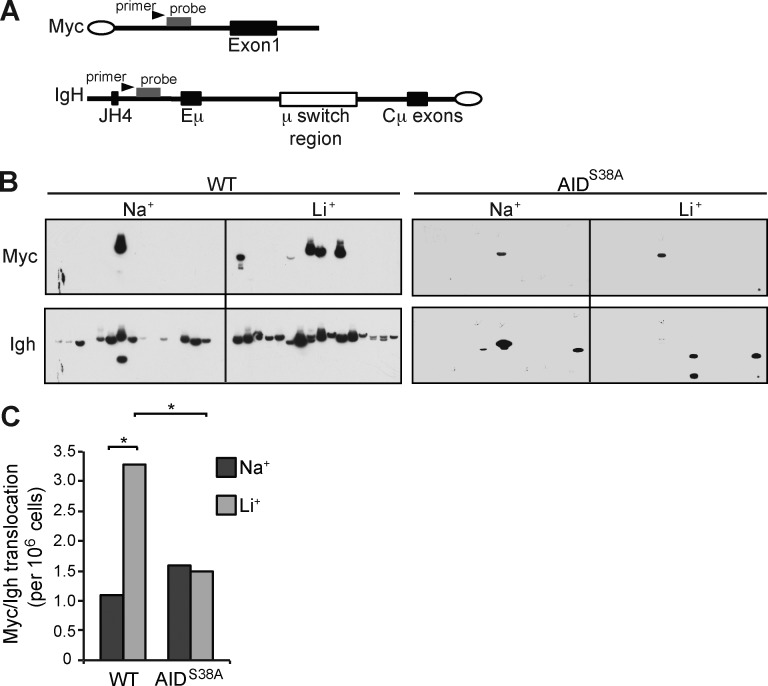
AID activity at off-target sites can induce DSB substrates for chromosome translocations. AID induces the oncogenic Myc/Igh translocation by causing DSBs at both Igh and Myc, with DSBs at Myc being rate limiting (Ramiro et al., 2004; Robbiani et al., 2008). The contribution of pS38 to off-target AID activity is not known. Therefore, we determined the effect of increased pS38 on the generation of Myc/Igh translocations. We used a previously described PCR/Southern blot assay to determine translocation frequency (Fig. 4 A). LiCl treatment of WT cells resulted in a significant threefold translocation increase compared with control (WT: NaCl control 7, and LiCl 21, translocations from 6.4 × 106 cells; Fig. 4, B and C). AIDS38A B cells displayed a translocation frequency similar to that of control WT cells irrespective of LiCl treatment (AIDS38A: NaCl control 19, and LiCl 18, translocations from 1.22 × 107 cells; Fig. 4 C). Interestingly AIDS38A B cells displayed a lower frequency of amplicons recognized by the Igh probe, which sequencing revealed to be internal switch region rearrangements (Fig. 4 A). We conclude that AID generates a background rate of translocations in a pS38-independent manner. However, supraphysiologic pS38 levels induce a significant translocation increase. Because DSBs at Myc are a limiting factor in translocation generation, this result is consistent with enhanced pS38 driving DSBs at Myc.
Figure 4.
AID Serine 38 phosphorylation promotes Myc/Igh translocations. (A) Schematic for the Myc/Igh translocation assay. PCR amplification primers are represented by black arrows and Southern probes by gray bars. Closed circles denote centromeric locations on the chromosomes. (B) Representative translocation assay Southern blots with Myc and Igh probes are displayed; each lane contains the DNA content of 1×105 genomes. B cells from WT or AIDS38A mice cultured with IL-4 and LPS were treated with 10 mM NaCl or LiCl for 12 h before analysis. (C) Total translocation frequency summary from n = 4 independent experiments. P-values (*, P < 0.01) determined using two-tailed Fisher’s exact test (WT Na+ vs. AIDS38A Na+ or Li+, both P > 0.5).
Phosphorylation promotes differential targeting of AID mutation activity
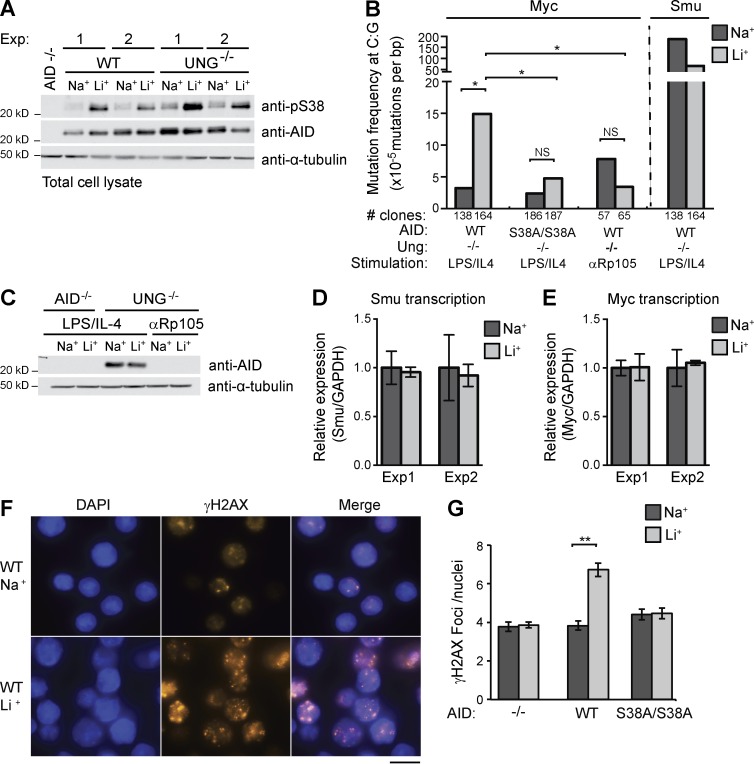
To determine whether pS38 regulates AID mutation activity at Myc, we analyzed the frequency of AID-induced point mutations. AID associates with Myc, but mutation is low because of high-fidelity repair by UNG or mismatch repair (Liu et al., 2008). Indeed, we found that Myc mutation frequency in WT B cells was not significantly higher than the PCR error rate regardless of treatment (not depicted). Unrepaired uracils accumulate in the absence of UNG, which results in C:G to T:A transition mutations when replicated. B cells deficient in UNG incur more numerous Myc mutations, predominantly at C:G residues (Liu et al., 2008; Yamane et al., 2011). Therefore, we analyzed Myc in B cells from Ung−/− mice and focused on mutation at C:G. LiCl induced pS38 to a similar extent in WT and Ung−/− B cells (Fig. 5 A). In Ung−/− cells, LiCl treatment resulted in a fivefold increase in mutation frequency compared with NaCl control, which was near background (Fig. 5 B and Table S1; Liu et al., 2008; McBride et al., 2008). This increase is statistically significant and occurred in the 12-h treatment span. Almost all the LiCl-induced mutations were at C:G sites and were transitions, and most were at AID hotspot (WRC) motifs (Table S1).
Figure 5.
Phosphorylation promotes differential targeting of AID mutation activity. (A) Anti-pS38, anti-AID, and anti–α-tubulin immunoblot of cell lysates from AID−/−, WT, or Ung−/− B cells activated for 72 h and treated with 10 mM LiCl (Li+) or control NaCl (Na+) for 12 h. Two independent experiments displayed. (B) Mutation frequency at C:G bases in Myc or Igh Smu. B cells from Ung−/− or Ung−/−/AIDS38A/S38A double-mutant mice were stimulated as indicated. Graph is summary of n = 5 independent experiments. Number of clones sequenced indicated. P-values (*, P < 0.05) were determined by a two-tailed t test assuming unequal variance (Myc: WT Na+ vs. Li+ and AIDS38A Na+ vs. Li+, both not significant [NS], P > 0.4, Smu: Na+ vs. Li+, P = 0.07). (C) Anti-AID and anti–α-tubulin immunoblot of lysates from AID−/− or UNG−/− B cells activated with either LPS/IL-4 or anti-Rp105. Representative of n = 2 experiments. (D and E) Igh Smu germline (D) and Myc (E) transcript level analysis. Levels of each transcript from treated B cells were analyzed by quantitative RT-PCR and quantified relative to Gapdh transcript levels. Control NaCl sample transcript levels arbitrarily set to 1; n = 2 experiments are displayed. Error bars are SE. (F) Representative image of WT B cell nuclei stained with DAPI (blue) and anti-γH2AX (red). Bar, 10 µm. (G) Mean number of foci in nuclei positive for γH2AX staining. Data representative of n = 3 experiments (error bars, SEM); **, P < 0.001; t test.
To ensure that the LiCl-induced mutations were caused by AID activity, we stimulated Ung−/− B cells with anti-Rp105, which induced proliferation but not AID expression (Fig. 5 C; Callén et al., 2007). In these cells, LiCl did not induce mutation higher than control levels (Fig. 5 B). To demonstrate the role of pS38 in LiCl-induced mutation, we analyzed B cells from Ung−/−/AIDS38A double-mutant mice. Myc mutation was not significantly different from background regardless of treatment (Fig. 5 B and Table S1). To boost the sensitivity and directly detect uracils in Myc, we amplified the same genomic DNA with Pfu-Cx, a mutant polymerase that can amplify uracil-containing DNA (Wang et al., 2017). Although we found a mutation frequency higher than background level, there was no difference between LiCl-treated and control NaCl-treated cells (Fig. S2). Strikingly, and in contrast to Myc, LiCl treatment resulted in a threefold mutation decrease at the switch µ (Smu) region (Fig. 5 B and Table S1), suggesting dynamic alterations in AID activity targeting.
To ensure that LiCl did not alter target transcription rates, we analyzed the relative levels of Smu region and Myc transcripts (Fig. 5, D and E) by quantitative RT-PCR and observed no change in transcript levels after LiCl treatment. We concluded that LiCl treatment results in differential AID-induced mutation, with an increase at the off-target Myc gene and no increase at the on-target Smu region. This is consistent with differential targeting of AID-induced recombination, where an increase in Myc translocation coincides with a decrease in CSR (Figs. 3 C and 4 C). Because DSBs at Myc are limiting in the formation of Myc/Ig translocations, even the reduced CSR in AIDS38A B cells should not be limiting. Our analysis of B cells from AIDS38A mice supports the conclusion that pS38 increased AID activity at Myc. To determine whether LiCl-induced pS38 increased mutations at other loci, we examined the Ly6e and Il4ra genes, both reported to be AID targets (Robbiani et al., 2009; Yamane et al., 2011). However, we did not see increased mutation (Table S1), suggesting that not all loci are affected by LiCl-induced pS38. To determine the extent of LiCl-induced AID activity genome-wide, we assessed DSB formation by γH2AX foci formation (Fig. 5 F). Although the percentage of cells with foci was slightly increased in WT LiCl-treated cells (WT: NaCl 68%, LiCl 86%), the number of foci in each γH2AX-positive-staining cell was significantly increased (WT: NaCl 3.8 vs. LiCl 6.7 foci/cell) This increase was AID and pS38 dependent, as it did not occur in AID−/− or AIDS38A cells (Fig. 5 G).
Concluding remarks
How AID mutator activity is targeted is a critical question related to lymphomagenesis. AID has promiscuous genomic occupancy, but the involvement of posttranslational modifications in regulating AID off-target activity has not been studied. We show that pS38 is a major contributor to differential mutation activity. AIDS38A B cell analysis suggests that pS38 is regulating AID physiological activity at the Ig but not the Myc locus. Our discovery that LiCl alters AID pS38 levels allowed us to examine the consequences of enhanced pS38 levels. We performed a variety of activity analyses comparing 10 mM LiCl treatment for 12 h (Figs. 2, 3, 4, and 5). Our results suggested that AID phosphorylation can dynamically target AID activity at different genomic locations and contribute to genome instability. Although our γH2AX foci analysis demonstrated that AID activity was increased at multiple loci, mutational analysis of Il4ra and Ly6e genes suggested that only a subset of AID targets may be affected.
The observed LiCl-induced AID activity might suggest an association of long-term lithium use with lymphoma; however, such a prediction is complicated. Although B cell tumors have been reported in long-term lithium users, significant lymphoma incidence has not. Furthermore, long-term lithium use is actually associated with an overall lower incidence of cancer (Martinsson et al., 2016). Lithium affects several pathways that affect B cell function and are potential therapy targets, such as GSK3 in multiple myeloma (Piazza et al., 2013). In this study, we focused on pS38 alterations during short-term LiCl treatment because longer treatment complicates the epigenetic and gene expression landscape. We targeted GSK3 because previous studies suggested that the pathway regulated AID activity in CSR (Omori et al., 2006; Thornton et al., 2016). Although lithium is a GSK3 inhibitor (IC50 2 mM), use of the specific GSK3 inhibitor CHIR99021 did not mimic the effects of lithium. Using CHX, we demonstrated that enhanced pS38 is a direct effect and not caused by gene expression alterations. Quite a number of signaling pathways, including PI3K/Akt, Wnt/B-Catenin, MEK/ERK, oxidative stress, PI/PKC, and cAMP, are affected by lithium (Fig. S1 D; Lenox and Wang, 2003; Chiu et al., 2013). The molecular mechanisms and spectra of pathways modulated by lithium are not completely understood. Altered signaling is a common feature of lymphomas and a consequence of some therapies, the potential of which to affect genome instability and tumor resistance through AID is only beginning to be realized (Compagno et al., 2017). The unexpected finding that a drug in clinical use can alter AID activity targeting suggests that a more comprehensive picture of the signaling pathways that impact AID phosphorylation is needed.
Materials and methods
Mice
Wild-type, Aicda−/− (Muramatsu et al., 2000), AicdaF/F (Pavri et al., 2010), Ung−/− (Endres et al., 2004), and AicdaS38A/S38A (McBride et al., 2008) were on C57BL6/J background. Animals were bred and housed at the accredited University of Texas MD Anderson Cancer Center animal facility. All work was approved by the Institutional Animal Care and Use Committee in accordance with the National Institutes of Health guidelines for use of live animals.
Cell culture and treatment
B cell isolation and culture have been previously described (McBride et al., 2006). Naive B cells were purified from mouse spleens by anti-CD43 bead (Miltenyi Biotec) depletion and cultured in LPS (25 µg/ml; Sigma-Aldrich) and IL-4 (5 ng/ml; Sigma-Aldrich) together or 2.5 µg/ml anti-Rp105/CD180 (BD Biosciences) for 72 h. CH12 cells (Nakamura et al., 1996) were stimulated with TGF-β (1 ng/ml; R&D Systems), IL-4, and either LPS (25 µg/ml) or anti-CD40 antibody (1 µg/ml; eBioscience) for 48 h. Treatments with CHIR99021 (Selleckchem), cycloheximide (Sigma-Aldrich), NaCl, LiCl, and LiAc (Sigma-Aldrich) were initiated at indicated times before culture harvest and analysis. All NaCl/LiCl treatment comparisons were between cells of the same cultures.
Flow cytometry
For Ig class switch assays, cell suspensions were stained with allophycocyanin-conjugated anti-IgG1 antibodies (BD Biosciences). Samples were acquired on a LSRFortessa (BD) and analyzed with FlowJo (Tree Star). Flow cytometric analysis of surface Ig expression was performed on day 3 of culture with scatter gating and propidium iodide staining to exclude dead cells. Means were obtained from triplicate cultures of six spleens in two independent experiments.
Chromosome translocation assay
The translocation assay has been previously described (Kovalchuk et al., 1997; Ramiro et al., 2004; Gazumyan et al., 2011). Naive B cells were cultured with LPS and IL-4 for 72 h, with LiCl treatment for the final 12 h. PCR reactions (105 cells per reaction) were performed with first-round PCR primers, 5′-ACTATGCTATGGACTACTGGGGTCAAG-3′ and 5′-GTGAAAACCGACTGTGGCCCTGGAA-3′, and second-round PCR primers, 5′-CCTCAGTCACCGTCTCCTCAGGTA-3′ and 5′-GTGGAGGTGTATGGGGTGTAGAC-3′, to amplify Myc/Igh translocations. Amplicons were confirmed as translocations with reactivity to both Southern blot probes Myc, 5′-GGACTGCGCAGGGAGACCTACAGGGG-3′, and Igh, 5′-GAGGGAGCCGGCTGAGAGAAGTTGGG-3′. Data are a summary of four independent experiments, and p-values were calculated by two-tailed Fisher’s exact test.
Cell lysis and chromatin fractionation
Total cell lysates were made by 30-min RIPA buffer with protease inhibitor (0.2 mM PMSF, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin, and 2 mM EGTA) and phosphatase inhibitor (25 mM NaF, 1 mM sodium orthovanadate, 50 mM disodium β-glycerolphosphate, and 10 mM sodium pyrophosphate) extraction and used for immunoprecipitation or direct Western analysis. Chromatin fractionation was a modification from Perfetti et al. (2015). Cells were lysed in EBC-1 buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5% NP-40, 1 mM EDTA, 1 mM DTT, 0.2 mM PMSF, 1 µg/ml aprotinin, 1 μg/ml leupeptin, 1 µg/ml pepstatin, 2 mM EGTA, 25 mM NaF, 1 mM sodium orthovanadate, 50 mM disodium β-glycerolphosphate, and 10 mM sodium pyrophosphate). Nuclear pellets were collected, washed in EBC-1, and resuspended in EBC-2 (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 5 mM CaCl2, 0.2 mM PMSF, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 2 mM EGTA, 25 mM NaF, 1 mM sodium orthovanadate, 50 mM disodium β-glycerolphosphate, and 10 mM sodium pyrophosphate) for 30 min. The insoluble chromatin was pelleted, washed with EBC-2, resuspended in RIPA buffer, sonicated, and analyzed as the chromatin-bound fraction.
Antibodies and protein analysis
The antibodies used in this study were anti-pS38 (McBride et al., 2006), anti-AID (30F12 from Cell Signaling Technology or McBride et al. [2004]), and anti–α-tubulin (Sigma-Aldrich). Anti-γH2AX (20E3), anti–histone H3 (D1H2), anti-CDC6 (C42F7), and phospho-substrate antibodies 9611, 2261, and 9621, all from Cell Signaling Technology. Anti-FLAG M2 agarose (Sigma-Aldrich) was used for Flag immunoprecipitation and eluted with Flag peptide. Immunoprecipitated AID or cell lysates (70 µg) were separated on 12% NuPAGE (Invitrogen), transferred to PVDF membrane, probed with anti-pS38 antibody, stripped, and probed with anti-AID and then anti–α-tubulin antibody. Band intensity was quantified with ImageJ, and the ratios of pS38 to AID or AID to H3 signals were calculated. Relative changes were normalized to control samples.
Mutation and DNA damage analysis in primary B cells
Mutations were analyzed in cultured B cells from Ung−/− or Ung−/−AicdaS38A/S38A double-mutant mice. B cells were isolated from five separate mice and cultured independently. A fragment from the upstream region of Igh Smu region, Myc, Ly6e, or Il4ra genes was amplified with PfuTurbo (Agilent) under the following cycling conditions: 95°C for 3 min, then 25 cycles of 95°C for 30 s, 54°C for Myc and 57°C for Smu for 15 s, 68°C for 2 min, and 72°C for 7 min. Each group was amplified in triplicate and cloned into pCR4-TOPO (Invitrogen), and individual colonies were sequenced. Statistical significance was determined by two-tailed Student’s t test assuming unequal variance. Primers used (Robbiani et al., 2009; Yamane et al., 2011) were Sμ: forward, 5′-GACCCAGGCTAAGAAGGCAATC-3′, and reverse, 5′-GCGGCCCGGCTCATTCCAGTTCATTACAG-3′; Myc: forward, 5′-TGGTCTTTCCCTGTGTTCTTTCTG-3′, and reverse, 5′-GACACCTCCCTTCTACACTCTAAACCG-3′; Ly6e: forward, 5′-CTTGGGTGCATTCAGCCTTTG-3′, and reverse, 5′-CCTGAGGAACACCTCCACAAAC-3′; and Il4ra: forward, 5′-GAGCCTGAACTCGCAGGTAG-3′, and reverse, 5′-CCCTCAGTCAGAGAGCACAG-3′.
γH2AX foci formation analysis was performed according to Hasham et al. (2010). Cultured cells were from two mice per experiment. Cells were formaldehyde fixed, permeabilized, blocked, and stained with DAPI, anti–phospho γH2AX antibody, and anti-rabbit secondary antibody. Fluorescence microscopy was preformed with a Zeiss Axiophot, 63× objective, and the mean number of foci per cell was calculated from 150 cells per group.
Quantitative PCR for Myc and germline transcripts
RNA from treated, cultured B cells was extracted with GenElute mammalian total RNA miniprep kit (Sigma-Aldrich), and cDNA was produced with Superscript II reverse transcription (Invitrogen) according to the manufacturer’s protocol. Quantitative PCR was performed using Brilliant III SYBR Green Q-PCR Master Mix (Agilent). Reactions were performed in triplicate from B cells of three mice of each genotype. MX3005P Q-PCR software (Stratagene) was used for analysis, and values were normalized to GAPDH. Primers used (Bothmer et al., 2013) were μGLT: forward, 5′-TAGTAAGCGAGGCTCTAAAAAGCA-3′, and reverse, 5′-AGAACAGTCCAGTGTAGGCAGTAGA-3′, and Myc: forward, 5′-TGGAACTTACAATCTGCGAGCCAG-3′, and reverse, 5′-TCGCTCTGCTGTTGCTGGTGATAG-3′.
Online supplemental material
Fig. S1 shows anti-pS38 antibody recognition of enhanced AID phosphorylation. Fig. S2 shows the mutation frequency of Myc in AIDS38A B cells using Pfu-Cx polymerase. Table S1 shows the mutation analysis of Myc and Igh Smu.
Supplementary Material
Acknowledgments
We would like to thank T. Honjo for Aicda−/− mice, R. Jaenisch for Ung−/− mice, and Joshua Plummer for his assistance with flow cytometry and microscopy.
We acknowledge the University of Texas M.D. Anderson Cancer Center (UTMDACC) Research Animal Support Facility (NIH CA16672), flow cytometry core, and molecular biology core. This research was supported by the Welch Foundation (G-1847), Three Strohm Sisters Family Foundation, and UTMDACC Center for Environmental and Molecular Carcinogenesis and Leukemia Specialized Program of Research Excellence (NIH CA100632). Y. Mu was supported by a fellowship from the UTMDACC Center for Cancer Epigenetics.
The authors declare no competing financial interests.
Author contributions: Y. Mu, M. Zelazowska, and K. McBride designed and performed experiments and analyzed data. Y. Mu and K. McBride wrote the manuscript. K. McBride supervised the project.
Footnotes
Abbreviations used:
- AID
- activation-induced cytidine deaminase
- CHX
- cycloheximide
- CSR
- class switch recombination
- DSB
- double-strand break
- IC50
- half-maximal inhibitory concentration
- SHM
- somatic hypermutation
- Smu
- switch µ
- UNG
- uracil DNA glycosylase
References
- Alt F.W., Zhang Y., Meng F.L., Guo C., and Schwer B.. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 152:417–429. 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U., Chaudhuri J., Alpert C., Dutt S., Ranganath S., Li G., Schrum J.P., Manis J.P., and Alt F.W.. 2005. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 438:508–511. 10.1038/nature04255 [DOI] [PubMed] [Google Scholar]
- Bothmer A., Rommel P.C., Gazumyan A., Polato F., Reczek C.R., Muellenbeck M.F., Schaetzlein S., Edelmann W., Chen P.L., Brosh R.M. Jr., et al. . 2013. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J. Exp. Med. 210:115–123. 10.1084/jem.20121975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callén E., Jankovic M., Difilippantonio S., Daniel J.A., Chen H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L., et al. . 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 130:63–75. 10.1016/j.cell.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Casellas R., Basu U., Yewdell W.T., Chaudhuri J., Robbiani D.F., and Di Noia J.M.. 2016. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat. Rev. Immunol. 16:164–176. 10.1038/nri.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V., Bortnick A., and Murre C.. 2015. AID targeting: Old mysteries and new challenges. Trends Immunol. 36:527–535. 10.1016/j.it.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji M., Unniraman S., McBride K.M., and Schatz D.G.. 2007. Role of activation-induced deaminase protein kinase A phosphorylation sites in Ig gene conversion and somatic hypermutation. J. Immunol. 179:5274–5280. 10.4049/jimmunol.179.8.5274 [DOI] [PubMed] [Google Scholar]
- Cheng H.L., Vuong B.Q., Basu U., Franklin A., Schwer B., Astarita J., Phan R.T., Datta A., Manis J., Alt F.W., and Chaudhuri J.. 2009. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc. Natl. Acad. Sci. USA. 106:2717–2722. 10.1073/pnas.0812304106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.T., Wang Z., Hunsberger J.G., and Chuang D.M.. 2013. Therapeutic potential of mood stabilizers lithium and valproic acid: Beyond bipolar disorder. Pharmacol. Rev. 65:105–142. 10.1124/pr.111.005512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M., Wang Q., Pighi C., Cheong T.C., Meng F.L., Poggio T., Yeap L.S., Karaca E., Blasco R.B., Langellotto F., et al. . 2017. Phosphatidylinositol 3-kinase δ blockade increases genomic instability in B cells. Nature. 542:489–493. 10.1038/nature21406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J.M., and Neuberger M.S.. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22. 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Endres M., Biniszkiewicz D., Sobol R.W., Harms C., Ahmadi M., Lipski A., Katchanov J., Mergenthaler P., Dirnagl U., Wilson S.H., et al. . 2004. Increased postischemic brain injury in mice deficient in uracil-DNA glycosylase. J. Clin. Invest. 113:1711–1721. 10.1172/JCI200420926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazumyan A., Timachova K., Yuen G., Siden E., Di Virgilio M., Woo E.M., Chait B.T., Reina San-Martin B., Nussenzweig M.C., and McBride K.M.. 2011. Amino-terminal phosphorylation of activation-induced cytidine deaminase suppresses c-myc/IgH translocation. Mol. Cell. Biol. 31:442–449. 10.1128/MCB.00349-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin F.K., Jamison K.R., and Ghaemi S.N.. 2007. Manic-Depressive Illness: Bipolar Disorders and Recurrent Depression. Oxford University Press, New York. 1262 pp. [Google Scholar]
- Hakim O., Resch W., Yamane A., Klein I., Kieffer-Kwon K.R., Jankovic M., Oliveira T., Bothmer A., Voss T.C., Ansarah-Sobrinho C., et al. . 2012. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 484:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasham M.G., Donghia N.M., Coffey E., Maynard J., Snow K.J., Ames J., Wilpan R.Y., He Y., King B.L., and Mills K.D.. 2010. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat. Immunol. 11:820–826. 10.1038/ni.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P.S., and Melton D.A.. 1996. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA. 93:8455–8459. 10.1073/pnas.93.16.8455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk A.L., Müller J.R., and Janz S.. 1997. Deletional remodeling of c-myc-deregulating chromosomal translocations. Oncogene. 15:2369–2377. 10.1038/sj.onc.1201409 [DOI] [PubMed] [Google Scholar]
- Le Q., and Maizels N.. 2015. Cell cycle regulates nuclear stability of AID and determines the cellular response to AID. PLoS Genet. 11:e1005411 10.1371/journal.pgen.1005411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenox R.H., and Wang L.. 2003. Molecular basis of lithium action: Integration of lithium-responsive signaling and gene expression networks. Mol. Psychiatry. 8:135–144. 10.1038/sj.mp.4001306 [DOI] [PubMed] [Google Scholar]
- Liu M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., and Schatz D.G.. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845. 10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Maddu N., and Raghavendra P.B.. 2015. Review of lithium effects on immune cells. Immunopharmacol. Immunotoxicol. 37:111–125. 10.3109/08923973.2014.998369 [DOI] [PubMed] [Google Scholar]
- Martinsson L., Westman J., Hällgren J., Ösby U., and Backlund L.. 2016. Lithium treatment and cancer incidence in bipolar disorder. Bipolar Disord. 18:33–40. 10.1111/bdi.12361 [DOI] [PubMed] [Google Scholar]
- Matthews A.J., Husain S., and Chaudhuri J.. 2014. Binding of AID to DNA does not correlate with mutator activity. J. Immunol. 193:252–257. 10.4049/jimmunol.1400433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K.M., Barreto V., Ramiro A.R., Stavropoulos P., and Nussenzweig M.C.. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244. 10.1084/jem.20040373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K.M., Gazumyan A., Woo E.M., Barreto V.M., Robbiani D.F., Chait B.T., and Nussenzweig M.C.. 2006. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc. Natl. Acad. Sci. USA. 103:8798–8803. 10.1073/pnas.0603272103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K.M., Gazumyan A., Woo E.M., Schwickert T.A., Chait B.T., and Nussenzweig M.C.. 2008. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J. Exp. Med. 205:2585–2594. 10.1084/jem.20081319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot S.P., and Di Noia J.M.. 2017. Molecular mechanisms of somatic hypermutation and class switch recombination. Adv. Immunol. 133:37–87. 10.1016/bs.ai.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., and Honjo T.. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Kondo S., Sugai M., Nazarea M., Imamura S., and Honjo T.. 1996. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int. Immunol. 8:193–201. 10.1093/intimm/8.2.193 [DOI] [PubMed] [Google Scholar]
- Omori S.A., Cato M.H., Anzelon-Mills A., Puri K.D., Shapiro-Shelef M., Calame K., and Rickert R.C.. 2006. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 25:545–557. 10.1016/j.immuni.2006.08.015 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Kitaura Y., Gu H., and Dalla-Favera R.. 2006. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc. Natl. Acad. Sci. USA. 103:395–400. 10.1073/pnas.0509969103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V., et al. . 2010. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 143:122–133. 10.1016/j.cell.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfetti M.T., Baughman B.M., Dickson B.M., Mu Y., Cui G., Mader P., Dong A., Norris J.L., Rothbart S.B., Strahl B.D., et al. . 2015. Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1. ACS Chem. Biol. 10:1072–1081. 10.1021/cb500956g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza F., Manni S., and Semenzato G.. 2013. Novel players in multiple myeloma pathogenesis: Role of protein kinases CK2 and GSK3. Leuk. Res. 37:221–227. 10.1016/j.leukres.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Eisenreich T., Difilippantonio S., Chen-Kiang S., Muramatsu M., Honjo T., Nussenzweig A., and Nussenzweig M.C.. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438. 10.1016/j.cell.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Ring D.B., Johnson K.W., Henriksen E.J., Nuss J.M., Goff D., Kinnick T.R., Ma S.T., Reeder J.W., Samuels I., Slabiak T., et al. . 2003. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 52:588–595. 10.2337/diabetes.52.3.588 [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., and Nussenzweig M.C.. 2013. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu. Rev. Pathol. 8:79–103. 10.1146/annurev-pathol-020712-164004 [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Bothmer A., Callen E., Reina-San-Martin B., Dorsett Y., Difilippantonio S., Bolland D.J., Chen H.T., Corcoran A.E., Nussenzweig A., and Nussenzweig M.C.. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 135:1028–1038. 10.1016/j.cell.2008.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Bunting S., Feldhahn N., Bothmer A., Camps J., Deroubaix S., McBride K.M., Klein I.A., Stone G., Eisenreich T.R., et al. . 2009. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell. 36:631–641. 10.1016/j.molcel.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryves W.J., and Harwood A.J.. 2001. Lithium inhibits glycogen synthase kinase-3 by competition for magnesium. Biochem. Biophys. Res. Commun. 280:720–725. 10.1006/bbrc.2000.4169 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E., and Schrader C.E.. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292. 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T.M., Delgado P., Chen L., Salas B., Krementsov D., Fernandez M., Vernia S., Davis R.J., Heimann R., Teuscher C., et al. . 2016. Inactivation of nuclear GSK3β by Ser(389) phosphorylation promotes lymphocyte fitness during DNA double-strand break response. Nat. Commun. 7:10553 10.1038/ncomms10553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong B.Q., Lee M., Kabir S., Irimia C., Macchiarulo S., McKnight G.S., and Chaudhuri J.. 2009. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat. Immunol. 10:420–426. 10.1038/ni.1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong B.Q., Herrick-Reynolds K., Vaidyanathan B., Pucella J.N., Ucher A.J., Donghia N.M., Gu X., Nicolas L., Nowak U., Rahman N., et al. . 2013. A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat. Immunol. 14:1183–1189. 10.1038/ni.2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Kieffer-Kwon K.R., Oliveira T.Y., Mayer C.T., Yao K., Pai J., Cao Z., Dose M., Casellas R., Jankovic M., et al. . 2017. The cell cycle restricts activation-induced cytidine deaminase activity to early G1. J. Exp. Med. 214:49–58. 10.1084/jem.20161649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A., Resch W., Kuo N., Kuchen S., Li Z., Sun H.W., Robbiani D.F., McBride K., Nussenzweig M.C., and Casellas R.. 2011. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol. 12:62–69. 10.1038/ni.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.