Abstract
Lipid biosynthesis within the chloroplast, or more generally plastids, was conventionally called “prokaryotic pathway,” which produces glycerolipids bearing C18 acids at the sn-1 position and C16 acids at the sn-2 position, as in cyanobacteria such as Anabaena and Synechocystis. This positional specificity is determined during the synthesis of phosphatidate, which is a precursor to diacylglycerol, the acceptor of galactose for the synthesis of galactolipids. The first acylation at sn-1 is catalyzed by glycerol-3-phosphate acyltransferase (GPAT or GPT), whereas the second acylation at sn-2 is performed by lysophosphatidate acyltransferase (LPAAT, AGPAT, or PlsC). Here we present comprehensive phylogenomic analysis of the origins of various acyltransferases involved in the synthesis of phosphatidate, as well as phosphatidate phosphatases in the chloroplasts. The results showed that the enzymes involved in the two steps of acylation in cyanobacteria and chloroplasts are entirely phylogenetically unrelated despite a previous report stating that the chloroplast LPAAT (ATS2) and cyanobacterial PlsC were sister groups. Phosphatidate phosphatases were separated into eukaryotic and prokaryotic clades, and the chloroplast enzymes were not of cyanobacterial origin, in contrast with another previous report. These results indicate that the lipid biosynthetic pathway in the chloroplasts or plastids did not originate from the cyanobacterial endosymbiont and is not “prokaryotic” in the context of endosymbiotic theory of plastid origin. This is another line of evidence for the discontinuity of plastids and cyanobacteria, which has been suggested in the glycolipid biosynthesis.
Keywords: acyltransferase, cyanobacteria, endosymbiosis, phosphatidate phosphatase, plastid
Introduction
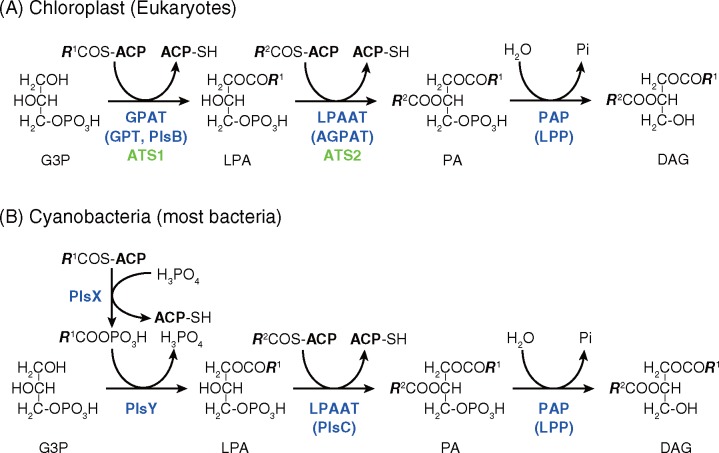
Glycerolipid biosynthesis in the chloroplast, or more generally plastids, starts by the synthesis of phosphatidate (PA) by two-step reactions (see fig. 1 for the pathways). The initial acylation of glycerol 3-phosphate (G3P) at its sn-1 position is catalyzed by glycerol 3-phosphate acyltransferase, which is called GPAT in plants and GPT in animals and yeasts. In the chloroplasts, it is called ATS1. The second acylation, namely, the acylation at the sn-2 position of lysophosphatidate (LPA) or 1-acyl-glycerol-3-phosphate, the product of the first acylation, is catalyzed by lysophosphatidate acyltransferase (LPAAT or LPAT in plants, ATS2 in chloroplasts) or 1-acyl-glycerol-3-phosphate acyltransferase (AGPAT in animals). Bacterial LPAAT is called PlsC. A part of PA will be used for the synthesis of phosphatidylglycerol, whereas most of PA will then be dephosphorylated by PA phosphatases, which are members of a large family of lipid phosphate phosphatases (LPPs). The product, diacylglycerol (DAG), will be used for the synthesis of galactolipids and sulfolipid in the chloroplast. There are two major galactolipids in the chloroplasts and cyanobacteria, namely, monogalactosyl diacylglycerol (MGDG) and digalactosyl diacylglycerol (DGDG). The specificity of the two acyltransferases determines the variety of molecular species of initial products of galactolipid synthesis within the chloroplast. The acyl groups may then be desaturated, exchanged by lipid retailoring (or remodeling) processes, or degraded to give the steady-state composition of lipid molecular species found in the actual biological membranes.
Fig. 1.
—Pathways of diacylglycerol synthesis in chloroplasts (A) and cyanobacteria (B). ACP, acyl carrier protein; PA, phosphatidic acid; PAP, phosphatidate phosphatase; Pi, inorganic phosphate. PlsB, PlsX, PlsY, ATS1, and ATS2 are gene names.
In plants and algae, fatty acid synthesis is performed predominantly in the chloroplast (or the plastids in the nongreen tissues of plants or the algae of different colors) (Guschina and Harwood 2006; Merchant et al. 2012; Hori et al. 2016; Li-Beisson et al. 2015; Bates 2016; Mori et al. 2016; Zienkiewicz et al. 2016). Mitochondria have a very minor activity of fatty acid synthesis. Unlike animals and fungi, no cytosolic fatty acid synthases are present in plants and algae (Archaeplastida). The fatty acids synthesized within the chloroplasts are either used for the synthesis of “prokaryotic” acyl lipids directly within the chloroplasts or transported to the cytosol for the subsequent synthesis of acyl lipids in the endoplasmic reticulum (ER). The acyl groups are desaturated and/or elongated there, and then a significant part of them return to the chloroplasts for the synthesis of “eukaryotic” galactolipids. Here the words “prokaryotic” and “eukaryotic” refer to the different biosynthetic origins of lipids. This naming was originally inspired by the fact that cyanobacterial lipids known at the time comprised 1-C18-2-C16 species and were thought to be similar to the molecular species of chloroplast lipids (Zepke et al. 1978; Roughan and Slack 1982; Sato and Murata 1982). This is not true for all cyanobacteria (e.g., Prochlorococcus marinus contains C30 and C28 MGDG, namely, C14/C16 and C14/C14 molecular species, respectively; Sato 2015), but the lipids of model cyanobacteria, such as Anabaena and Synechocystis, fit to the simplified view. The words “prokaryotic” and “eukaryotic” are also used symbolically to mean that chloroplasts originated from prokaryotic (or cyanobacterial) endosymbiont (note that Mereschkowsky 1905 was cited in Introduction of Zepke et al. 1978 in p. 157), whereas the ER represents the eukaryotic site of metabolism. This notion was then extended to the two pathways of lipid biosynthesis: The “prokaryotic pathway” produces PA within the chloroplasts, whereas the “eukaryotic pathway” involves the complex traffic of fatty acids from the chloroplast to the ER and then to the chloroplast again (Roughan and Slack 1982; Somerville and Browse 1991; Li-Beisson et al. 2015; Zienkiewicz et al. 2016), although these pathway names were not used initially. In a primitive red alga Cyanidioschyzon merolae, the synthesis of a single molecule of MGDG required cooperation of both pathways, and this was named specifically “cooperative pathway” (Sato and Moriyama 2007).
The synthesis of specific molecular species of lipids in the two pathways has been explained by the distinct specificities of the two acyltransferases producing PA in the chloroplasts and the ER, respectively. In the current understandings, the names of the two pathways lost meanings for specific molecular species, but they just indicate two distinct pathways for the synthesis of plastid lipids. Several papers were published on the evolution of biosynthesis of plastid lipids, especially galactolipids (Petroutsos et al. 2014; Hori et al. 2016). Unfortunately, no phylogenetic analysis was provided in the study of Petroutsos et al. (2014), which presented comparison of enzymes in different organisms. Hori et al. (2016) presented phylogenetic trees, but the number of taxa analyzed was quite limited, and the bootstrap values of the major branches were significantly low for deducing evolutionary narratives. We need a more rigorous and convincing phylogenetic analysis to understand the origin of lipid biosynthetic pathways in plastids.
The origin of acyltransferases in the plastids has been believed naïvely to be cyanobacterial enzymes. A report on the phylogenetic analysis of LPAAT suggested that the chloroplast LPAAT (ATS2) originated from the cyanobacterial PlsC (Körbes et al. 2016), but the very limited taxon sampling made us question this conclusion. The PA phosphatases of the chloroplast were characterized as prokaryotic in a previous report (Nakamura et al. 2007). However, the presented phylogenetic tree was not convincing to identify the chloroplast PA phosphatases as prokaryotic. We therefore exploited the custom-made phylogenomic clusters obtained by the Gclust software (Sato 2009) for the comprehensive analysis of acyltransferases and PA phosphatases to find the origins of these enzymes in detail. An advantage of using Gclust is its capability to explicitly detect the N-terminal extensions or transit peptides of eukaryotic proteins having homologs in prokaryotes. We already exploited this advantage of Gclust clusters and succeeded in analyzing the vast protein families such as the PsbP proteins (Sato 2010) and the protoporphyrin-IX oxidases (Kobayashi et al. 2014), the cyanobacterial enzymes in the galactolipid synthesis (Sato and Awai 2016), as well as the enzymes in the chloroplast peptidoglycan synthesis (Sato and Takano 2017).
In this study, we analyzed the phylogeny of the predominant acyltransferases and PA phosphatases in about 170 organisms encompassing prokaryotes and eukaryotes including both photosynthetic and nonphotosynthetic organisms. The results do not support the previous statements that the chloroplast enzymes originated from a cyanobacterial endosymbiont. The chloroplast enzymes are basically part of conserved eukaryotic enzymes, but could have diverse, distant probable origins: Namely, the first acylation enzyme is related to chlamydial enzymes, and the second acylation enzyme is related to the homologs in green bacteria, whereas the phosphatase originated from eukaryotic enzymes. We will also discuss the implications of this finding in the light of the theory of the endosymbiotic origin of plastids.
Data and Methods
Sequence Data
We used the preformed homolog clusters in the Gclust database (Sato 2009) as available in the web site http://gclust.c.u-tokyo.ac.jp (data set 2012_42) to find clusters including acyltransferases using known names of proteins. This data set contained protein sequences of various photosynthetic organisms (cyanobacteria, photosynthetic bacteria, algae, and plants) as well as nonphotosynthetic organisms (various bacteria, Archaea, fungi, protists, and animals). The sequences obtained from data set 2012_42 are marked by the cluster number, which appears at the top of each protein ID. The original sources of the sequences are described in the web site. We did not detect acyltransferases in Archaea, maybe because they do not have acyl lipids. To find homologs of red algae and other organisms, we also used data set Gclust2017R6, which was an enhanced version of data set Gclust2016R (Sato and Moriyama 2017), which contained six red algae and three diatoms as well as small number of other phyla. Additional sequences in data set Gclust2017R6 were two Paulinella chromatophore genomes (Nowack et al. 2008; Lhee et al. 2017), Micromonas commoda RCC299 (MicpuN3v2_GeneCatalog_proteins_20160404, Joint Genome Institute Genome Portal, Worden et al. 2009), Micromonas pusilla CCMP1545 (MicpuC3v2_GeneCatalog_proteins_20160125, Joint Genome Institute Genome Portal, van Baren et al. 2016), Cyanophora paradoxa (protein sequences 022111) from the Cyanophora Genome Project (http://cyanophora.rutgers.edu/cyanophora/home.php, Price et al. 2012), and Pseudo-nitzschia multiseries CLN-47 (Psemu1_GeneCatalog_proteins_20111011, Joint Genome Institute Genome Portal). To obtain all available homologs of ATS1, we performed a BLASTP search for the nr database of National Center for Biotechnology Information (as of September 29, 2017) and retrieved 100 homologs for each of the 28 proteins of the phylogenetic tree in figure 3B. After removing redundancy, about 350 sequences remained, comprising only Chlamydiae bacteria, plants, and algae. Phylogenetic tree was constructed by the maximum likelihood (ML) method. The validity of homology clustering by Gclust was evaluated by calculating an identity score for each alignment by the “simtbl” command (enhanced in version 159.44) of SISEQ (Sato 2000).
Fig. 3.
—Phylogenetic tree of PlsX (A), PlsY (C), and ATS1(B, D). (A) and (C) are collapsed BI trees. The values on each branch indicate a posterior probability of BI/a confidence value of ML. (D) is a collapsed ML tree using all available sequences of ATS1 homologs. The alignment files are available as supplementary data S2–S5, Supplementary Material online. The uncollapsed BI and ML trees for (A) are available as supplementary figures S1 and S2, Supplementary Material online, respectively. The uncollapsed BI and ML trees for (C) are available as supplementary figures S3 and S4, Supplementary Material online, respectively. The uncollapsed ML tree for (D) is available as supplementary figure S5, Supplementary Material online. A tentative position of the root is indicated in (D).
Phylogenetic Analysis
In the attempt of all-inclusive phylogenetic reconstruction, we used all acyltransferase sequences as a whole to construct a single large tree. The methods of alignment and phylogenetic reconstruction were essentially identical to those used in a previous paper on the phylogeny of peptidoglycan synthesizing enzymes (Sato and Takano 2017): Protein sequences were aligned by the software Muscle version 3.8.31 (Edgar 2004). The alignment was visualized by the software Clustal X version 2 (Larkin et al. 2007). Distant sequences were removed, and ill-aligned N- and C-terminal ends were trimmed by the “getclu” command of the software SISEQ (Sato 2000). Only the sites having gaps in less than 20% of the total sequences were used for the calculation (“gap 0.2” option of “getclu” command).
To validate alignments, the HoT (heads or tails) method was applied (Landan and Graur 2007) by using the “rev” option of the “getclu” command of SISEQ. In this procedure, a multifasta file was reversed to obtain a “tails” fasta file, which was then aligned by muscle, and then reversed again. After applying the “gap 0.2” command, the heads alignment and the tails alignment were compared. For this purpose, the order of sequences within the alignment was sorted (the “sort_name” option of the “getclu” command), and then all the site data were retrieved as a single file (the “site_dump” option). After checking the divergent removal of sites by the gap removal process, the two files were treated as two sets of transposed sequences, and the differences were counted on the file prepared by the “save column scores” command of Clustal X. Only 1.9% of residues were inconsistent in the two alignments (heads and tails) of ATS1. In ATS2 and PlsC, about 90% of residues were consistent, and only 7 sites within the total 167 sites (except 4 sites that were differently removed in the heads and tails processing) were significantly different. This did not affect significantly the tails tree of ATS2 and PlsC (supplementary fig. S13, Supplementary Material online). Essential similarity of the tails tree with the heads tree (fig. 4) suggested the validity of the sequence alignment and phylogenetic analysis. For LPP, the conservation of heads and tails was inferior. We performed GUIDANCE2 analysis (Sela et al. 2015) and selected 50% of the total 165 sites to prepare an ML tree (supplementary fig. S14, Supplementary Material online), which was essentially similar to the tree in figure 6, although some mixing of taxa occurred due to lower resolution resulting from smaller number of sites. The results supported the validity of the alignment and phylogenetic tree of LPP.
Fig. 4.
—Phylogenetic tree of ATS2 and PlsC. This is a collapsed BI tree of all members of Clusters 488 and 4242 with red algal sequences. The values on each branch indicate (a posterior probability of BI)/(a confidence value of ML). This tree was tentatively rooted according to the MAD method (Tria et al. 2017). Uncollapsed BI and ML trees are available as supplementary figures S6 and S7, Supplementary Material online. The alignment is available as supplementary data S6, Supplementary Material online. An alternative tree (BI/ML) without new sequences from Gclust2017R6 is available as supplementary figure S8, Supplementary Material online.
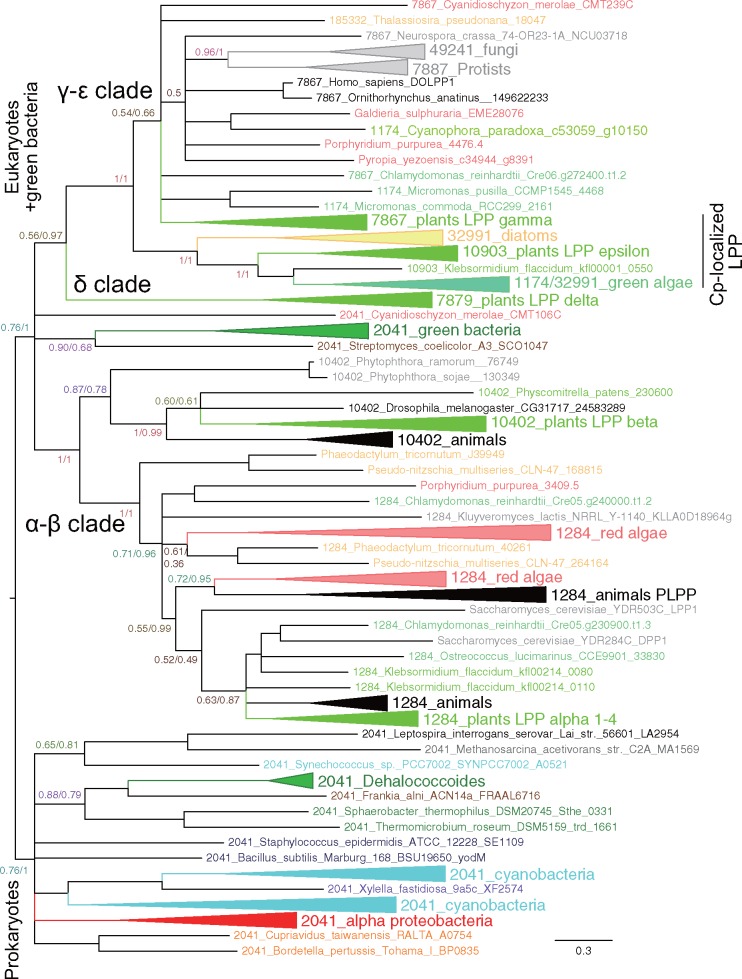
Fig. 6.
—Phylogenetic tree of phosphatidate phosphatases. This is a collapsed BI tree, with a posterior probability of BI/a confidence value of ML at each major branch. This tree was rooted according to the MAD method and is consistent with prokaryote/eukaryote dichotomy. The alignment is available as supplementary data S10, Supplementary Material online. Uncollapsed BI and ML trees are available as supplementary figures S11 and S12, Supplementary Material online, respectively.
Initial phylogenetic tree was constructed by the ML method using the software PhyML version 3 (Guindon et al. 2010) (options were: -d aa –m LG –s BEST –b -5). Then, removal of distant sequences, trimming of both ends, realignment, and ML calculation were repeated (about ten times) to obtain a reasonably reproducible tree. The sequences in Cluster 33000 were very long, and therefore, only the homologous region was identified and used in the alignment with all other sequences. In the initial analysis, PlsX and PlsY were also included with all other acyltransferases, but inclusion of these distant sequences prevented construction of reliable trees: the positions of various clusters changed with trials. That is why PlsX and PlsY were not included in the comprehensive phylogenetic reconstruction, but they were analyzed individually.
According to the result of all-inclusive phylogenetic tree, we selected closely related clusters, which were then analyzed individually. In this case, Bayesian inference (BI) analysis was also performed using the software MrBayes version 3.2.6 (Ronquist et al. 2012). LG model was used in both PhyML and MrBayes calculations. Other parameters in MrBayes were rates = invgamma (in some cases, gamma), ratepr = variable, and ngen = 2,000,000 (up to 50,000,000). samplefreq and burnin were appropriately set depending on the value of ngen. The burnin values were set to remove a half of the generated trees. All the calculation was performed in the Linux and MacPro workstations in the laboratory or in the supercomputer in the Human Genome Center, the University of Tokyo. Graphical representation of the final trees was prepared by the software FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/), followed by decoration by Adobe Illustrator version CS6.
The position of the root in some important trees was estimated using the R script of the MAD software (Tria et al. 2017).
In the main text, we show mainly collapsed versions of phylogenetic trees for simplicity in many cases. The full versions of both BI and ML trees are presented in supplementary figures, Supplementary Material online. The multiple alignments that were used for the phylogenetic calculations (after removing ill-aligned N- and C-terminal ends and gap sites) are also available as supplementary data, Supplementary Material online.
Results
All-Inclusive, Global Analysis of Acyltransferases
We first collected various homologs of known acyltransferases from the Gclust database (data set Gclust2012_42) containing all proteins of 169 organisms. As we collected sequences, we learned that some of the enzymes involved in the first acylation and the second acylation are homologs. It was also true that the plant LPAATs and GPATs were separated into several different subclusters (table 1). We also included sequences from recently sequenced red algal genomes (data set Gclust2017R6). The clusters of Gclust were “protein clusters” but not “domain clusters.” Gclust is similar to other clustering tools such as OrthoMCL (http://orthomcl.org/orthomcl/), but specialized for comparing organellar and bacterial homologs (Sato 2009). Homologous proteins can be present in different clusters depending on additional domains or short sequence motifs. In the phylogenomic analysis that we attempted, all these homologs had to be assembled. The situation is shown in table 1. Each of the Gclust clusters contained homologs that were characterized by a mean identity score, ranging from about 0.25 to 0.45. The identity score was higher in some small clusters. As expected, large clusters had lower identity scores. The values were roughly similar to or higher than those calculated for protein or domain clusters in the public databases (lower rows of table 1). We collected as many clusters as possible for acyltransferases and constructed a large multiple alignment (supplementary data S1a, Supplementary Material online). Weak homology was detected throughout the entire acyltransferases (identity score = 0.240 ± 0.130). Related clusters of LPPs were also assembled. The identity score of each group of sequences for a tree construction (acyltransferases and phosphatases) ranged from 0.23 to 0.40, which justified that the groups were indeed homologous, and relevant for constructing phylogenetic trees.
Table 1.
Sequence Similarity Within the Gclust Clusters and the Alignments for Trees
| Tree Name | Figure # | Gclust2012 |
Gclust2017R6 |
Merge | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster # | # of seq | E-value | Identity | Cluster # | # of seq | E-value | Identity | Identity | ||
| ATS1 | 3B | 7661 | 26 | 1.00E-80 | 0.452 ± 0.131 | 2193 | 17 | 1.00E-45 | 0.360 ± 0.103 | 0.405 ± 0.117 |
| PlsX | 3A | 1241 | 117 | 1.00E-31 | 0.405 ± 0.131 | not used | n/a | |||
| PlsY | 3C | 937 | 141 | 1.00E-22 | 0.420 ± 0.112 | not used | n/a | |||
| ATS2 & PlsC | 4 | 488 | 190 | 1.00E-10 | 0.250 ± 0.100 | 1090 | 26 | 1.00E-50 | 0.319 ± 0.098 | 0.270 ± 0.098 |
| 5855 | 7 | 1.00E-50 | 0.487 ± 0.084 | |||||||
| 4242 | 42 | 1.00E-28 | 0.420 ± 0.113 | 7159 | 6 | 1.00E-45 | 0.427 ± 0.082 | |||
| LPP | 6 | 1284 | 113 | 1.00E-60 | 0.453 ± 0.175 | 1174 | 25 | 1.00E-12 | 0.252 ± 0.070 | 0.231 ± 0.115 |
| 2041 | 76 | 1.00E-16 | 0.260 ± 0.104 | |||||||
| 7867 | 25 | 1.00E-35 | 0.433 ± 0.177 | |||||||
| 7879 | 25 | 1.00E-80 | 0.545 ± 0.190 | |||||||
| 10402 | 19 | 1.00E-50 | 0.348 ± 0.143 | |||||||
| 10903 | 18 | 1.00E-50 | 0.519 ± 0.186 | |||||||
| 32991 | 5 | 1.00E-45 | 0.339 ± 0.137 | |||||||
| 49241 | 3 | 1.00E-60 | 0.545 ± 0.046 | |||||||
| GPT4 | S8 | 5749 | 33 | 1.00E-60 | 0.388 ± 0.184 | 838 | 30 | 1.00E-50 | 0.246 ± 0.091 | 0.260 ± 0.113 |
| 6695 | 29 | 1.00E-45 | 0.247 ± 0.129 | |||||||
| 16298 | 12 | 1.00E-25 | 0.639 ± 0.314 | |||||||
| LPAAT7 | 5 | 4244 | 42 | 1.00E-99 | 0.405 ± 0.207 | 1066 | 26 | 1.00E-70 | 0.243 ± 0.063 | 0.302 ± 0.132 |
| 6202 | 31 | 1.00E-50 | 0.392 ± 0.181 | |||||||
| LPAAT9 | S7 | 5766 | 33 | 1.00E-70 | 0.400 ± 0.195 | 1492 | 21 | 1.00E-70 | 0.355 ± 0.106 | 0.333 ± 0.162 |
| 7815 | 25 | 1.00E-45 | 0.334 ± 0.171 | |||||||
| Reference clusters for comparison (acyltransferases) | ||||||||||
| NCBI CPDF | cd07984 | 100 | n/a | 0.176 ± 0.043 | ||||||
| cd07989 | 100 | n/a | 0.173 ± 0.050 | |||||||
| InterPro | IPR004552 | 100 | n/a | 0.371 ± 0.255 | ||||||
| 100 | n/a | 0.264 ± 0.122 | ||||||||
| 1000 | n/a | 0.278 ± 0.108 | ||||||||
| Pfam | PF01553 | 100 | n/a | 0.156 ± 0.056 | ||||||
| 100 | n/a | 0.150 ± 0.050 | ||||||||
Note.—The original clusters from Gclust2012 and Gclust2017R6 were individually aligned. The average identity ± standard deviation was calculated over all sequence pairs within each cluster. “Merge” indicates the data for the actual sequence alignments used for the phylogenetic tree construction. Comparable data of homologous sequence family were obtained from NCBI Cluster of Protein Domain, InterPro, and Pfam, and the average identity score was calculated in a similar way. “E-value” is the threshold of E-value. “n/a” indicates not applicable. For Reference clusters, 100 or 1000 sequences were arbitrarily taken from the original data and analyzed.
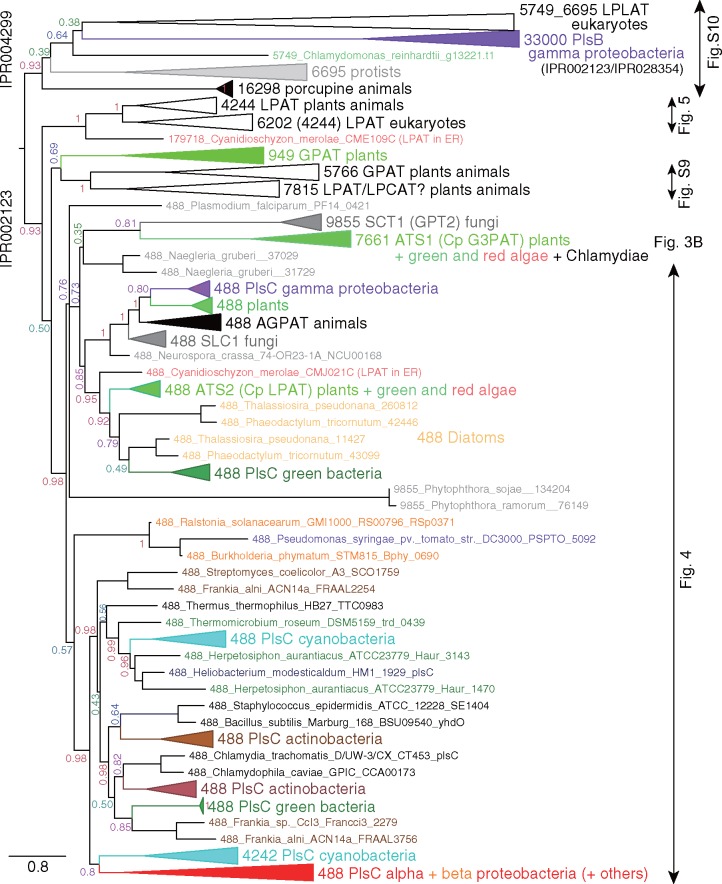
Figure 2 shows a global phylogenetic tree showing the overview of the acyltransferases. There were two major branches containing different InterPro motifs, IPR002123 and IPR004299 (see the web site of European Bioinformatics Institute: http://www.ebi.ac.uk/), which were supported by a high confidence level. The IPR004299 motif included mostly lysophospholipid acyltransferases (LPLAT). The bacterial PlsB, such as the one known in Escherichia coli, is a very large enzyme, and this type of acyltransferase was only present in the gamma-proteobacteria (the Cluster 33000). This cluster was located within the group containing the IPR004299 motif, but the reliability of the branching was not high. In fact, the position of this cluster changed with different sets of clusters and, therefore, was no longer studied.
Fig. 2.
—Phylogenetic tree of all acyltransferases showing global classification of acyltransferases. This is a collapsed ML tree. The value on each major branch indicates a confidence value. The upper branch contains sequences having InterPro motif IPR004299 (membrane-bound acyltransferase), whereas the lower branch contains sequences having IPR002123 (acyltransferase). The original tree file is available as supplementary data S1a, Supplementary Material online. Phyla are consistently color-coded according to the color scheme in the FigTree software throughout this paper (the original RGB intensities are shown in each parenthesis although the actual figures are printed in the CMYK mode): Green (or Spring) (0, 255, 0), land plants; Sea Foam (0, 255, 128), green algae; Salmon (255, 102, 102), red algae; Cantaloupe (255, 204, 102), diatoms; Red (or Maraschino) (255, 0, 0), alpha-proteobacteria; Tangerine (255, 128, 0), beta-proteobacteria; Grape (128, 0, 255), gamma-proteobacteria; Cyan (or Turquoise) (0, 255, 255), cyanobacteria; Clover (0, 128, 0), green bacteria; Midnight (0, 0, 128), Firmicutes; Aluminum (153, 153, 153), Archaea (very rare occurrence); Mocha (128, 64, 0), actinobacteria; Black (0, 0, 0), other bacteria and fungi (very rare occurrence).
In the clade having the IPR002123 motif, a single major cluster (Cluster 488) contained various LPAATs, such as the bacterial PlsC, the chloroplast ATS2, and the animal AGPAT. The GPAT clusters, namely, the Cluster 9855 including the fungal GPT and the Cluster 7661 including the chloroplast ATS1 and its chlamydial homologs, were found closely related to the Cluster 488, although the confidence level of the branching was not high. Various eukaryotic GPATs (Clusters 949, 5766, and 7815) and LPAATs (Clusters 4244 and 6202) were also present in the large group containing IPR002123. Because some major branches had low values of confidence, we decided to perform phylogenetic analysis for smaller clades as shown below. The full phylogenetic tree of figure 2, too large to be presented as a single figure, can be visualized by the FigTree software with the tree file supplied in supplementary data S1b, Supplementary Material online.
Enzymes Involved in the First Acylation in the Chloroplasts and Cyanobacteria
Figure 3 summarizes the phylogenetic trees of enzymes involved in the first acylation, namely, G3P acyltransferases. In bacteria, this step consists of two reactions: PlsX catalyzes the synthesis of acyl phosphate, whereas PlsY transfers this acyl group to G3P (fig. 1). In the original report (Lu et al. 2006), PlsX and PlsY were discovered in pathogenic Gram-positive bacteria, but most bacteria, including cyanobacteria, contain both PlsX and PlsY (fig. 3A and C). The chromatophore genomes of the two species of Paulinella (Nowack et al. 2008; Lhee et al. 2017) also encode plsX and plsY genes. This is consistent with the idea that the chromatophores of Paulinella originate from a recent endosymbiosis, which is distinct from the plastid endosymbiosis. None of the plants and algae contained these genes.
The acylation of G3P is catalyzed by ATS1 in the chloroplasts (fig. 3B). Curiously, the genomes of two chlamydial species (we used only two representative chlamydial species in the Gclust database) also encode homologs of the ATS1 gene named plsB, although they also encode plsX but not plsY. No homologs of chloroplast ATS1 are present in bacteria except these chlamydial sequences within the Gclust databases. To complement the results, we obtained all available homologs from the nr database of NCBI. We found homologs in only Chlamydiae bacteria, plants and algae. Some sequences of Apicomplexa were also found, but they have a long N-terminal domain, and might not be orthologs. The phylogenetic tree of all these sequences (fig. 3D) was consistent with the tree in figure 3B. The branching of diatom sequences (shown in yellowish orange) was different in figure 3B and D, but the reliability of the branching was 1.0 for both the Chlamydiae clade and the plants/algae clade. According to the rooting with the MAD software (Tria et al. 2017) as shown in figure 3D, we can imagine that Chlamydiae acquired ATS2 from algae, not vice versa. This point should be studied further. These results indicate that the G3P acylation is catalyzed by completely different enzymes in the chloroplasts and cyanobacteria. The original alignments of PlsX, ATS1, and PlsY are provided as supplementary data S2–S4, Supplementary Material online, respectively. The detailed phylogenetic trees of PlsX are provided as supplementary figures S1 (BI method) and S2 (ML method), Supplementary Material online, whereas the trees of PlsY are found in supplementary figures S3 (BI method) and S4 (ML method), Supplementary Material online. Uncollapsed ML tree of figure 3D is shown in supplementary figure S5, Supplementary Material online.
Enzymes Involved in the Second Acylation in the Chloroplasts and Cyanobacteria
Figure 4 shows a phylogenetic tree of all members of Clusters 488 and 4242. Each cyanobacterium contains two PlsC, one in Cluster 488 and the other in Cluster 4242. Cluster 4242 is not very different from Cluster 488 in overall alignment (supplementary data S6, Supplementary Material online: note that both N- and C-terminal ends were removed), but two short additional sequences are inserted. For example, a member of Cluster 4242, Sll1752 of Synechocystis sp. PCC 6803, contains two insertions: AGRGVTG after the position 39 (between ILLSLA and RDLRFM) in the alignment (note that this is a de-gapped alignment) and CKQNPNT from the position 119 (between IAMEV and DIKVI). Another copy of PlsC in Synechocystis sp. PCC 6803, Sll1848, is a member of Cluster 488. In the phylogenetic tree, the chloroplast homologs of bacterial PlsC, named ATS2, were clearly separated from the two cyanobacterial PlsC homologs. The clade containing enzymes of mostly cyanobacteria, actinobacteria, Firmicutes, and chlamydia was supported by a high value of confidence. The values shown in figure 4 was 0.96/– for BI/ML, but close examination of the original ML tree (supplementary fig. S7, Supplementary Material online) indicated that the clade containing the two cyanobacterial clades and various bacteria (actinobacteria, chlamydia, green bacteria, or Firmicutes) was also supported by the value 1.0. On the other hand, the clade containing all the eukaryotes and some bacteria was supported by high confidence (0.98/1 for BI/ML) in figure 4, although some differences in internal branching were noted in BI and ML trees. Figure 4 was rooted using the MAD software, and this position of the root seemed to be reasonable. These results indicate that the two cyanobacterial clades and the plants/algae clade were clearly separated as distant clades, despite small differences in the position of some bacterial sequences with respect to the cyanobacterial clades.
Arabidopsis thaliana has only a single ATS2, whereas some other plants such as rice contains another enzyme, which was closely related to the PlsC of gamma-proteobacteria (fig. 4 and see supplementary figs. S6 and S7, Supplementary Material online for details). These enzymes were predicted to be targeted to mitochondria. Animal AGPAT and fungal SLC1 (these are microsomal enzymes) were their sister groups. The chloroplast ATS2 of plants and algae (including the red algae and the glaucophyte) formed a single clade, which was sister to the clade of green bacterial PlsC, which was also sister to a small clade of diatom homologs. We noted, however, that the initial tree constructed with the Gclust 2012_42 data set, namely, before adding the enzymes of diatoms, red algae, and Paulinella (see Data and Methods), the green bacterial clade was located entirely outside the eukaryotic clade (supplementary fig. S8, Supplementary Material online). Addition of sequences of diatoms, secondary endosymbionts, could have modified the branching pattern. The rooting using the MAD method was somewhat different in the BI tree and the ML tree (indicated as an alternative root), but the estimated positions of the root in figure 4 and supplementary figure S8, Supplementary Material online, were always between the clade containing the cyanobacterial sequences and the clade containing the eukaryotic sequences. The support values of the clade consisting of green bacteria, eukaryotes, and gamma-proteobacteria were high in both figure 4 and supplementary figure S8, Supplementary Material online, indicating a close relationship of the green bacterial sequences with the eukaryotic sequences. The presence of some gamma-proteobacterial sequences (only enterobacteria) in a position close to the eukaryotic sequences is difficult to explain, but they could be acquired from the eukaryotes. These results did not agree with the previous claim that chloroplast ATS2 originates from cyanobacterial PlsC (Körbes et al. 2016). Chloroplast ATS2 (and eukaryotic homologs) might have originated from the green bacterial PlsC, but we need further studies.
Acyltransferases in the ER
Acyltransferases are also present in the ER. Although these enzymes are not primary focus of this study, we suspected if some of these originate from cyanobacteria. Many eukaryotic GPAT enzymes are found in Cluster 5766 (supplementary fig. S9, Supplementary Material online: alignment is in supplementary data S8, Supplementary Material online). They were divided into the clades of plants/algae and animals. This cluster was sisters to the putative LPAT or LPCAT in Cluster 7815. No cyanobacterial or prokaryote sequences were present in these clusters.
The enzymes of LPA acylation were also found in Clusters 4244 (including plant LPAT2/3) and 6202 (including plant LPAT4/5) (fig. 5: alignment is in supplementary data S7, Supplementary Material online). Enzymes of plants and animals were present in these two clusters, whereas fungal CST26 (in Cluster 4244) was related to Cluster 6202 of animals and plants. Red algal and glaucophyte enzymes were found only in the clade related to Cluster 4244, but curiously, they were not sister to the plant LPAT2/3. Note that the human enzyme in Cluster 6202 is annotated as lysocardiolipin acyltransferase. The three human enzymes in Cluster 4244 are annotated as AGPAT. No cyanobacterial or prokaryote homologs were found in these clusters.
Fig. 5.
—Phylogenetic tree of ER-localized LPAT/AGPAT/LCLAT group. This is a BI tree, with a posterior probability of BI/a confidence value of ML at each major branch. The alignment is available as supplementary data S7, Supplementary Material online.
The phylogenetic tree of enzymes having the InterPro motif IPR004299 is shown in supplementary figure S10 (alignment in supplementary data S9), Supplementary Material online. They included Clusters 5749 (membrane-bound acyl transferases or LPLAT), 6695 (mostly protist enzymes), and 16298 (protein-serine O-palmitoyltransferase: porcupine or PORCN). The red algal and diatom enzymes were not clustered with the plant and green algal enzymes, but with the protist enzymes. One of the two glaucophyte enzymes was found in the animal clade, and the other was related to the plant and green algal enzymes. These results showed that there is no acyltransferase of cyanobacterial origin in the entire lipid biosynthetic pathways in plants and algae.
Phylogenetic Analysis of PA Phosphatases
PA phosphatases are very diverse enzymes found in both prokaryotes and eukaryotes. They are dispersed in many clusters in the Gclust database (data set Gclust2012_42): We found Clusters 1284, 7867, 7879, 10402, 10903, 32991, and 49241, as well as some singletons for eukaryotic LPPs. We also found Cluster 2041 for various prokaryotic and some algal enzymes. In addition, we have collected homologs from the data set Gclust2017R6 for red algal, glaucophyte, and diatom sequences (Cluster 1174). All these sequences were homologs as evidenced by the average identity values in table 1. Figure 6 shows a simplified (collapsed) phylogenetic tree of all these enzymes (alignment is in supplementary data S10, Supplementary Material online). It is clear that they were simply separated into eukaryotic and prokaryotic clades, except one of the two clades of green bacterial enzymes, which was found within the eukaryotic clade. This dichotomy was consistent with the rooting with the MAD software. The confidence values of this major dichotomy were not high in BI analysis, because the position of this green bacterial clade was variable depending on calculations. Apart from such ambiguity, the separation of prokaryotic enzymes and eukaryotic enzymes was nearly complete. This was also supported by the tree constructed from reliable sites estimated by the GUIDANCE2 software (supplementary fig. S14, Supplementary Material online). This situation is very different from the phylogenetic tree presented in the work of Nakamura et al. (2007), stating that chloroplast enzymes were prokaryotic and possibly originated from cyanobacteria. This difference could be a result of large differences in taxon sampling and methods of phylogenetic reconstruction.
In the present tree of PA phosphatases, eukaryotic clades consisted of the α–β subclade and δ–γ–ɛ subclade. The latter was subdivided into the δ group and the γ–ɛ group. Except for the δ group, each group consisted of both plant and animal/fungal enzymes, suggesting an ancient origin of these five groups. The chloroplast-localized isozymes, LPPγ (AT5G03080), LPPɛ1 (AT3G50920), and LPPɛ2 (AT5G66450), were found within the eukaryotic γ–ɛ subclade. If we divide all the enzymes into (prokaryotic clade plus δ–γ–ɛ subclade) and α–β subclade (see the ML tree in supplementary fig. S12, Supplementary Material online), this corresponds roughly to the phylogenetic tree shown in the work of Nakamura et al. (2007). Even in this case, the presence of both plant and animal/fungal enzymes within each group, α, β, γ, and ɛ, was not compatible with the idea that the chloroplast enzymes originated from a cyanobacterial endosymbiont.
Discussion
Non-Cyanobacterial Nature of the “Prokaryotic Pathway”
The pathway of lipid biosynthesis within the chloroplast has been called “prokaryotic pathway” since the 1980s. This notion evoked that the lipid biosynthetic capacity of the chloroplast was acquired from a cyanobacterial endosymbiont. Galactolipids such as MGDG and DGDG are common basic constituents of the thylakoid membranes of chloroplasts and cyanobacteria. It is now established that these galactolipids are synthesized by entirely different, independent pathways in chloroplasts and cyanobacteria (Sato and Murata 1982; Awai et al. 2006, 2007; Sakurai et al. 2007; Awai et al. 2014; Sato and Awai 2016). This study adds further evidence for the differences in the pathways of lipid biosynthesis in the chloroplasts and the cyanobacteria.
DAG, a direct precursor for the synthesis of galactolipids, is produced by dephosphorylation of PA. As summarized in figure 1, the acylation of G3P, the initial step of PA synthesis, is catalyzed by entirely different pathways in cyanobacteria and chloroplasts. Cyanobacteria use PlsX and PlsY, whereas chloroplasts use ATS1, an enzyme unique to photosynthetic eukaryotes, but probably related to chlamydial PlsB. A large family of LPAAT enzymes are involved in the second acylation step in bacteria and eukaryotes: Cyanobacteria use two types of PlsC enzymes, as in many other bacteria (having one or two copies of PlsC), whereas chloroplasts use ATS2, which is distinct from the cyanobacterial PlsC within the LPAAT family. The closest prokaryotic relative of the chloroplast ATS2 is the PlsC of green bacteria, but the results of supplementary figure S8, Supplementary Material online, rather suggested that all of the eukaryotic ATS2/LPAAT enzymes are distinct from the prokaryotic enzymes from the origin of eukaryotes (i.e., not a product of horizontal gene transfer).
It appears that the dephosphorylation of PA is catalyzed by various different enzymes in bacteria and chloroplasts. Enzymatic activity of a cyanobacterial PA phosphatase (Sll0545) demonstrated in yeast transformants (Nakamura et al. 2007). In plants, various phosphatases belonging to the LPP family have been known to act as PA phosphatases (Nakamura et al. 2007). Among them, LPPγ, LPPɛ1, and LPPɛ2 were considered as the chloroplast-localized PA phosphatases. PA phosphatases might be quite variable depending on organisms. In the red alga C. merolae, CMT106C was targeted to the cytosol, whereas all other PA phosphatases identified by homology were localized to the ER (Mori et al. 2016). No PA phosphatase has yet been localized to the plastid in C. merolae.
It is also noted that not all bacterial species appeared in the LPP tree (fig. 6), even though we used a comprehensive comparative genome database including many prokaryotes from representative phyla. This could indicate that many bacteria do not possess this enzyme (DAG is not necessary for bacterial phospholipid biosynthesis in general). However, some enzymes must still have escaped identification. For example, we did not identify PA phosphatase homologs in various strains of P. marinus, a vast group of marine cyanobacteria, which also contain galactolipids as major membrane lipids (see, e.g., Sato 2015). Obviously, various phosphatases are encoded by the genomes of Prochlorococcus, and we will have to specify putative PA phosphatase in the future. To this end, we detected putative phosphatases (HAD-like phosphatases like eukaryotic PAH1/2) in the chromatophore genome of Paulinella, which have homologs in cyanobacteria and other prokaryotes (Clusters 4706 and 7215 in Gclust2017R6), and could be involved in the dephosphorylation of PA or phosphatidylglycerol phosphate. This is a strategy that we took in identifying the epimerase gene mgdE in cyanobacteria (Awai et al. 2014). The chromatophore genome of Paulinella conserves various essential genes for the synthesis of lipid synthesis within the chromatophore, and this fact provides a convenient method of identifying cyanobacterial homologs. In any case, these are prokaryotic enzymes and different from eukaryotic LPPs. Based on these results, we can conclude that the pathway of DAG synthesis in the chloroplast did not originate from the corresponding pathway in cyanobacteria.
Taken together, the pathway of lipid biosynthesis starting from G3P until DGDG is fundamentally different in cyanobacteria and chloroplasts except DgdA in C. merolae that we identified before (Awai et al. 2007; Sakurai et al. 2007). In other words, the “prokaryotic pathway” of chloroplast lipid biosynthesis did not originate from cyanobacteria and was not a result of endosymbiosis, even if the chloroplasts are supposed to be a descendant of a cyanobacterial endosymbiont (Archibald 2015; Sato 2016).
Revision of Previous Phylogenetic Analyses
Körbes et al. (2016) showed a close relationship between the cyanobacterial PlsC and the chloroplast ATS2. They used only the PlsC domain (125 amino acid residues in the final alignment) of a very limited number of related taxa (208 sequences of 44 species) in constructing a phylogenetic tree of all LPAATs (namely, prokaryotic, eukaryotic, and organellar ones). This could account for the sister relationship of cyanobacterial and chloroplast LPAATs, which they showed in the paper. In contrast, we used a fairly large number of bacterial PlsC in inferring phylogenetic relationship between the plant and cyanobacterial homologs. The region of sequence analysis included the domain used by Körbes et al. (2016), but was larger (325 and 171 amino acid residues, before and after gap removal, respectively). We subdivided the phylogenetic analysis into closely related sequences according to the initial classification (fig. 2). The fact that the selection of homologs was based on the rational clustering by the Gclust made it easy to construct these subdivided phylogenetic trees. As a result, we found many taxa between the clade of cyanobacterial PlsC and the clade of chloroplast ATS2 (figs. 2 and 4), which clearly indicated that the chloroplast ATS2 did not originate from cyanobacterial PlsC.
The phylogenetic tree of PA phosphatases in this study is an alternative to the previously published one (Nakamura et al. 2007). We already discussed this discrepancy with the authors of the paper, who are, in fact, collaborators in our different projects. We are sure that they will agree with us that we will no longer be able to use the notation “prokaryotic” (this appeared in the title of the paper) for the chloroplast-localized phosphatases, which were identified as eukaryotic as described above.
Non-Cyanobacterial Origin of Chloroplast Membranes
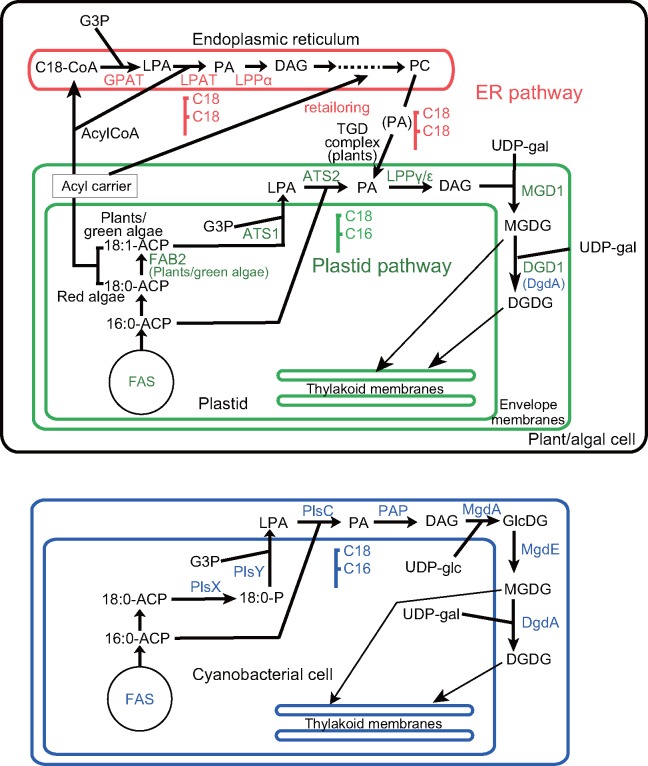
The results of this study suggest that the pathway of galactolipid synthesis is different in the chloroplasts and cyanobacteria (fig. 7). None of the enzymes involved in the synthesis of MGDG and DGDG are shared by cyanobacteria (blue) and chloroplasts (green). Similar 1-C18-2-C16 species of these lipids are synthesized in both cyanobacteria and chloroplasts, but this is not a result of the working of “prokaryotic pathway” in the chloroplasts. We might call the plastid-localized pathway of lipid biosynthesis “plastid pathway” instead of “prokaryotic pathway.” As a corollary, it might be better to call the pathway in the ER “ER pathway” rather than “eukaryotic pathway.” Nevertheless, because this is a complex pathway involving both ER and plastid envelope, the eukaryotic pathway could still be used. The pathway localized in the ER membrane (red) is essentially shared by all eukaryotes, including plants, algae, protists, fungi, and animals. The “eukaryotic lipid” such as the 1-C18-2-C18 species might be specific to the plants and algae, because the animals and fungi typically contain 1-saturated-2-unsaturated species. In marine red algae, the 1-C20-2-C20 species of MGDG and DGDG (Sato et al. 2017) are typically produced as a result of transport of precursor lipid from the ER to the chloroplasts. Therefore, the meaning of “eukaryotic lipid” is ambiguous. Only the pathway is important.
Fig. 7.
—Comparison of lipid biosynthetic pathways in plastids (top) and cyanobacteria (bottom). All enzymes are color-coded: blue, cyanobacterial or prokaryotic enzymes; green, plastid-localized enzymes; red, ER-localized enzymes. The latter two categories of enzymes are eukaryotic enzymes (of eukaryotic origin), except the TGD complex components that are of cyanobacterial origin (see, e.g., Hori et al. 2016). In plants and green algae, stearoyl-ACP desaturase (FAB2) catalyzes the conversion of 18:0 to 18:1 within the plastid, whereas in red algae, this enzyme is not present, and the final product of fatty acid synthesis within the plastid is 18:0. The C18 acids are transported out of the plastids and used to synthesize the 1-C18-2-C18 lipids in the ER. Desaturation occurs on these lipids, but this is not shown explicitly. The PA or DAG portion of phosphatidylcholine comes back to the plastids and used for the synthesis of the galactolipids. The pathway via ER is a very simplified sketch, because it is not the main topic of this study. Within the plastids (plastid pathway), the 1-C18-2-C16 lipids (sometimes called “prokaryotic lipids”) are synthesized by ATS1 and ATS2, which are not of cyanobacterial origin. MGDG is synthesized by galactosylation of DAG catalyzed by MGD1, and DGDG by galactosylation of MGDG catalyzed by DGD1. In the red algae, C. merolae and Galdieria sulphuraria, the last step is catalyzed by DgdA (or Ycf82), a cyanobacterial enzyme encoded by the plastid genome. Other red algae possess DGD1 but not DgdA. In cyanobacteria, MGDG is synthesized by the epimerization (catalyzed by MgdE) of GlcDG, which is synthesized by the glucosylation of DAG catalyzed by MgdA. DgdA is the sole enzyme catalyzing the production of DGDG in cyanobacteria. ACP, acyl carrier protein; CoA, coenzyme A; FAS, fatty acid synthase; GlcDG, monoglucosyl diacylglycerol; UDP-glc, uridine diphosphate glucose; UDP-gal, uridine diphosphate galactose.
In many popular drawings of the theory of the endosymbiotic origin of chloroplasts (e.g., fig. 1 of Archibald 2015), a cyanobacterial cell is engulfed by a host cell, giving a symbiotic cell. The resultant chloroplast possesses two envelope membranes and numerous thylakoid membranes, which are typically colored like the cytoplasmic membranes and thylakoid membranes of the cyanobacterial progenitor (such as fig. 2 of Petroutsos et al. 2014). Such picturesque view on the endosymbiotic origin of chloroplasts might be misleading, because all membranes of chloroplasts contain galactolipids that are synthesized by the enzymes that did not originate from cyanobacteria. If the membranes of chloroplasts did not originate from cyanobacteria, then it will be difficult to draw the chloroplasts, in such figures, as the cyanobacteria living in a plant cell. In other words, the popularized visual image of the endosymbiosis has been collapsed. As proposed already (Sato 2001; Moriyama and Sato 2014; Sato 2016), there is a clear discontinuity in the genomic machinery of chloroplasts, although the chloroplast genome itself is likely a descendent of some ancestral cyanobacterial genome.
We could imagine that ATS1 originated from Chlamydiae, because Chlamydia and Parachlamydia (and some related species) were the only bacterial phyla that have ATS1 homologs (fig. 3D). However, a tentative estimation of the root of the tree suggested that Chlamydiae acquired ATS1 from the algae, but not the other way. It is interesting to note that an involvement of a chlamydial cell during the establishment of plastid endosymbiosis was proposed (Ball et al. 2011; Cenci et al. 2017). These authors hypothesized that the host cell starved by the damage of pathogen infection could be rescued by photosynthesis by cyanobacterial endosymbiont. A detailed analysis of glycogen-related metabolic enzymes concluded, however, that there is no compelling evidence that chlamydia played an important role in plastid establishment (Domman et al. 2015). A critical argument was also given by Dagan et al. (2013), stating that “only one endosymbiosis with many lateral gene transfers.”
It should also be noted that several papers described that a significant part of the chloroplast proteome consists of enzymes that originated from various prokaryotes other than cyanobacteria (Qiu et al. 2013; Ku et al. 2015). This is essentially in line with the results of this study. The analyses in the two papers, however, relied on the comparison of homologs. As shown in this study, a single reaction could be catalyzed by nonhomologous enzymes of different origins. To find such evolutionary replacement, it will be necessary to analyze each reaction one by one.
Another paper (Pittis and Gabaldón 2016) presented evidence that the mitochondrial proteins of alpha-proteobacterial origin showed shorter phylogenetic distance to their closest prokaryotic relatives, compared with proteins of different prokaryotic origin, stating that the horizontal gene transfer resulting in the latter occurred before the mitochondrial acquisition. We have examined whether this method can be useful in our study. This can only be applied to ATS2 (fig. 4), because the chloroplast ATS1 is present mostly in eukaryotic plants and algae and that the chloroplast LPP is derived from eukaryotic LPP. We calculated the stem length of eukaryotic lineage (the length between the separation from the green bacterial lineage and the diversification of green, red, and glaucophyte lineages) relative to the median of the distances of green lineage (rather than the whole eukaryotic lineage as used in the original). The use of the distance of green lineage for normalization was justified, because red and glaucophyte lineages are not always available for comparison, depending on the proteins/genes to be compared. The results were 0.31 and 0.26 for ATS2 and plastid rRNA, respectively. The time of acquisition of ATS2 might be similar to the time of the primary endosymbiosis. However, an alternative tree (supplementary fig. S8, Supplementary Material online) suggested that ATS2 was a member of eukaryotic LPAATs. In this case, ATS2 was not a product of horizontal gene transfer. Obviously, we will have to work more on this subject by examining a more plausible phylogenetic tree and assessing the evolutionary rates of the different lineages. This will be the next project involving a larger number of chloroplast proteins.
In this analysis, only diatoms were used as the secondary endosymbionts, which are thought to be the results of endosymbiosis of a red algal cell. The ancestry of red algal sequences with respect to the diatom sequences is expected for the plastid proteins of endosymbiont origin, but this is not always the case in most analyses in this study. Inclusion of diatom sequences might have modified the branching patterns in some analyses. We need further studies to obtain a reliable phylogenetic position of diatom sequences.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Dr Yuki Nakamura (Academia Sinica, Taiwan) for discussion on the phylogeny of PA phosphatases. We also thank the Associate Editor for helpful suggestions. Some of the very large calculations were performed in the supercomputers of the Human Genome Center, University of Tokyo. This work was supported in part by JSPS KAKENHI (15K12433, 17H03715 to N.S.) from Japan Society for the Promotion of Science, and a grant, Core Research for Evolutional Science and Technology, from Japan Science and Technology Agency to N.S. and K.A. N.S. and K.A. conceived the research. N.S. conducted data analysis. K.A. worked on the characterization of PlsX, PlsY, and PlsC. N.S. and K.A. wrote the manuscript.
Literature Cited
- Archibald JM. 2015. Endosymbiosis and eukaryotic cell evolution. Curr Biol. 2519:R911–R921. [DOI] [PubMed] [Google Scholar]
- Awai K, Kakimoto T, Awai C, Kaneko T, Nakamura Y.. 2006. Comparative genomic analysis revealed a gene for monoglucosyl diacylglycerol synthase, an enzyme for photosynthetic membrane lipid synthesis in cyanobacteria. Plant Physiol. 1413:1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awai K, Ohta H, Sato N.. 2014. Oxygenic photosynthesis without galactolipids. Proc Natl Acad Sci U S A. 11137:13571–13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awai K, Watanabe H, Benning C, Nishida I.. 2007. Digalactosyldiacylglycerol is required for better photosynthetic growth of Synechocystis sp. PCC6803 under phosphate limitation. Plant Cell Physiol. 4811:1517–1523.http://dx.doi.org/10.1093/pcp/pcm134 [DOI] [PubMed] [Google Scholar]
- Ball S, Colleoni C, Cenci U, Raj JN, Tirtiaux C.. 2011. The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J Exp Bot. 626:1775–1801.http://dx.doi.org/10.1093/jxb/erq411 [DOI] [PubMed] [Google Scholar]
- Bates PD. 2016. Understanding the control of the acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim Biophys Acta 1861(9B):1214–1225. [DOI] [PubMed] [Google Scholar]
- Cenci U, et al. 2017. Biotic host-pathogen interactions as major drivers of plastid endosymbiosis. Trends Plant Sci. 224:316–328.http://dx.doi.org/10.1016/j.tplants.2016.12.007 [DOI] [PubMed] [Google Scholar]
- Dagan T, et al. 2013. Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol. 51:31–44.http://dx.doi.org/10.1093/gbe/evs117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domman D, Horn M, Embley TM, Williams TA.. 2015. Plastid establishment did not require a chlamydial partner. Nat Commun. 6:6421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 325:1792–1797.http://dx.doi.org/10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 593:307–321.http://dx.doi.org/10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Guschina IA, Harwood JL.. 2006. Lipids and lipid metabolism in eukaryotic algae. Prog Lipid Res. 452:160–186.http://dx.doi.org/10.1016/j.plipres.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Hori K, et al. 2016. Tangled evolutionary processes with commonality and diversity in plastidial glycolipid synthesis in photosynthetic organisms. Biochim Biophys Acta 1861(9B):1294–1308. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Masuda T, Tajima N, Wada H, Sato N.. 2014. Molecular phylogeny and intricate evolutionary history of the three isofunctional enzymes involved in the oxidation of Protoporphyrinogen IX. Genome Biol Evol. 68:2141–2155.http://dx.doi.org/10.1093/gbe/evu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körbes AP, Kulcheski FR, Margis R, Margis-Pinheiro M, Turchetto-Zolet AC.. 2016. Molecular evolution of the lysophosphatidic acid acyltransferase (LPAAT) gene family. Mol Phylogenet Evol. 96:55–69. [DOI] [PubMed] [Google Scholar]
- Ku C, et al. 2015. Endosymbiotic gene transfer from prokaryotic pangenomes: inherited chimerism in eukaryotes. Proc Natl Acad Sci U S A. 11233:10139–10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landan G, Graur D.. 2007. Heads or tails: a simple reliability check for multiple sequence alignments. Mol Biol Evol. 246:1380–1383.http://dx.doi.org/10.1093/molbev/msm060 [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 2321:2947–2948.http://dx.doi.org/10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lhee D, et al. 2017. Diversity of the photosynthetic Paulinella species, with the description of Paulinella micropora sp. nov. and the chromatophore genome sequence for strain KR01. Protist 1682:155–170.http://dx.doi.org/10.1016/j.protis.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Beisson F, Riekhof W.. 2015. Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 823:504–522.http://dx.doi.org/10.1111/tpj.12787 [DOI] [PubMed] [Google Scholar]
- Lu YJ, et al. 2006. Acyl-phosphates initiate membrane phospholipid synthesis in Gram-positive pathogens. Mol Cell 235:765–772.http://dx.doi.org/10.1016/j.molcel.2006.06.030 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J.. 2012. TAG, You’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr Opin Biotechnol. 233:352–363. [DOI] [PubMed] [Google Scholar]
- Mereschkowsky C. 1905. Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol Centralblatt 2518:593–604. [Google Scholar]
- Mori N, Moriyama T, Toyoshima M, Sato N.. 2016. Construction of global acyl lipid metabolic map by comparative genomics and subcellular localization analysis in the red alga Cyanidioschyzon merolae. Front Plant Sci. 7:958.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T, Sato N.. 2014. Enzymes involved in organellar DNA replication in photosynthetic eukaryotes. Front Plant Sci. 5:480.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Tsuchiya M, Ohta H.. 2007. Plastidic phosphatidic acid phosphatases identified in a distinct subfamily of lipid phosphate phosphatases with prokaryotic origin. J Biol Chem. 28239:29013–19021. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M, Glöckner G.. 2008. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 186:410–418. [DOI] [PubMed] [Google Scholar]
- Petroutsos D, et al. 2014. Evolution of galactoglycerolipid biosynthetic pathways—from cyanobacteria to primary plastids and from primary to secondary plastids. Prog Lipid Res. 54:68–85. [DOI] [PubMed] [Google Scholar]
- Pittis AA, Gabaldón T.. 2016. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 5317592:101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DC, et al. 2012. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 3356070:843–847.http://dx.doi.org/10.1126/science.1213561 [DOI] [PubMed] [Google Scholar]
- Qiu H, et al. 2013. Assessing the bacterial contribution to the plastid proteome. Trends Plant Sci. 1812:680–687.http://dx.doi.org/10.1016/j.tplants.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 613:539–542.http://dx.doi.org/10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan PG, Slack CR.. 1982. Cellular organization of glycerolipid metabolism. Annu Rev Plant Physiol. 331:97–132.http://dx.doi.org/10.1146/annurev.pp.33.060182.000525 [Google Scholar]
- Sakurai I, Mizusawa N, Wada H, Sato N.. 2007. Digalactosyldiacylglycerol is required for stabilization of the oxygen-evolving complex in photosystem II. Plant Physiol. 1454:1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N. 2000. SISEQ: manipulation of multiple sequence and large database files for common platforms. Bioinformatics 162:180–181.http://dx.doi.org/10.1093/bioinformatics/16.2.180 [DOI] [PubMed] [Google Scholar]
- Sato N. 2001. Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci. 64:151–156. [DOI] [PubMed] [Google Scholar]
- Sato N. 2009. Gclust: trans-kingdom classification of proteins using automatic individual threshold setting. Bioinformatics 255:599–605.http://dx.doi.org/10.1093/bioinformatics/btp047 [DOI] [PubMed] [Google Scholar]
- Sato N. 2010. Phylogenomic and structural modeling analyses of the PsbP superfamily reveal multiple small segment additions in the evolution of photosystem II-associated PsbP protein in green plants. Mol Phylogenet Evol. 561:176–186.http://dx.doi.org/10.1016/j.ympev.2009.11.021 [DOI] [PubMed] [Google Scholar]
- Sato N. 2015. Is monoglucosyl diacylglycerol a precursor to monogalactosyl diacylglycerol in all cyanobacteria? Plant Cell Physiol. 5610:1890–1899. [DOI] [PubMed] [Google Scholar]
- Sato N. 2016. Conservation versus discontinuity in the genealogy of cyanobacteria and plastids: fantasy and reality of the endosymbiogenesis theory of plastid origin. Endocytobiosis Cell Res. 273:33–36. [Google Scholar]
- Sato N, Awai K.. 2016. Diversity in biosynthetic pathways of galactolipids in the light of endosymbiotic origin of chloroplasts. Front Plant Sci. 7:117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Moriyama T.. 2007. Genomic and biochemical analysis of lipid biosynthesis in the unicellular rhodophyte Cyanidioschyzon merolae: lack of a plastidic desaturation pathway results in the coupled pathway of galactolipid synthesis. Eukaryot Cell 66:1006–1017.http://dx.doi.org/10.1128/EC.00393-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Moriyama T.. 2017. Chapter 18. Photosynthesis In: Kuroiwa T, Miyagishima S, Matsunaga S, Sato N, Nozaki H, Tanaka K, Misumi O, editors. Cyanidioschyzon merolae: a new model eukaryote for cell and organelle biology. Tokyo: Springer; (in press). [Google Scholar]
- Sato N, Moriyama T, Mori N, Toyoshima M.. 2017. Lipid metabolism and potentials of biofuel and high added-value oil production in red algae. World J Microbiol Biotechnol. 334:74..http://dx.doi.org/10.1007/s11274-017-2236-3 [DOI] [PubMed] [Google Scholar]
- Sato N, Murata N.. 1982. Lipid biosynthesis in the blue-green alga, Anabaena variabilis: I. Lipid classes. Biochim Biophys Acta 7103:271–278.http://dx.doi.org/10.1016/0005-2760(82)90109-6 [Google Scholar]
- Sato N, Takano H.. 2017. Diverse origins of enzymes involved in the biosynthesis of chloroplast peptidoglycan. J Plant Res. 1304:635–645. [DOI] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T.. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43(W1):W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Browse J.. 1991. Plant lipids: metabolism, mutants, and membranes. Science 2525002:80–87.http://dx.doi.org/10.1126/science.252.5002.80 [DOI] [PubMed] [Google Scholar]
- Tria FDK, Landan G, Dagan T.. 2017. Phylogenetic rooting using minimal ancestor deviation. Nat Ecol Evol. 1:0193.. [DOI] [PubMed] [Google Scholar]
- van Baren, et al. 2016. Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants. BMC Genomics 17:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, et al. 2009. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 3245924:268–272. [DOI] [PubMed] [Google Scholar]
- Zepke HD, Heinz E, Radunz A, Linscheid M, Pesch R.. 1978. Combination and positional distribution of fatty acids in lipid from blue-green algae. Arch Microbiol. 1192:157–162. [Google Scholar]
- Zienkiewicz K, Du ZY, Ma W, Vollheyde K, Benning C.. 2016. Stress-induced neutral lipid biosynthesis in microalgae—molecular, cellular and physiological insights. Biochim Biophys Acta 1861(9B):1269–1281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.