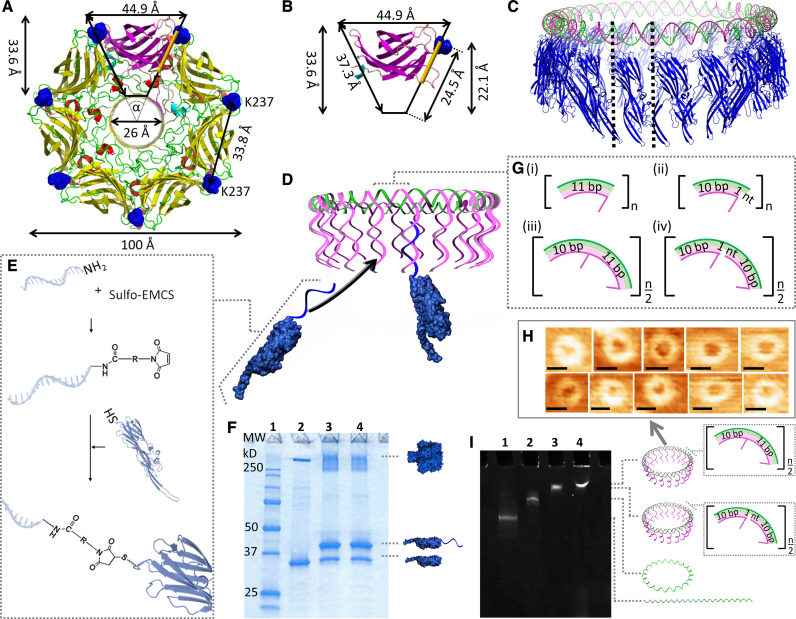
Figure 1.
Design of the DNA/αHL20 hybrid pore. Top view of (A), the overall structure of the wild-type αHL pore, and (B), the β-sandwich domain of a single protomer. The residue K237 is highlighted in blue. The protein pore can be simplified as a ring of 7 triangles with an angle, α, of ∼51.43°. The positions for possible monomer-monomer interactions within the cap domain in the region are highlighted in orange. (C) Comparison of the dimensions of a 220-bp long DNA ring (green, magenta) and the circular arrangement of 20 αHL monomers (blue). (D) Illustration of the assembly of the DNA/αHL hybrid structure. The DNA ring is composed of a closed single strand (the ‘scaffold strand’, green) that includes segments complementary to the 5′ end of the DNA protruding strands. K237C-αHL mutants that are modified with an oligonucleotide which is complementary to the 3′ end of these DNA strands can be arranged along the resulting DNA structure. (E) Schematic representation of the nucleic acid modification of a single αHL cys-mutant through a chemical crosslinker. (F) SDS-polyacrylamide gel showing unheated samples of unmodified (lane 2) and ssDNA-modified (lanes 3 and 4) K237C-αHL mutants. Lane 1 contains a protein standard; lanes 3 and 4 represent two different reactions under the same conditions. (G) Design of the repetitive sequence segments of the DNA template based on either 11 bp per one helical turn (i,ii), or 21 bp per two helical turns (iii, iv). (H) AFM images of DNA structures based on 21 bp per helical turn. Scale bars = 27 nm. (I) MgCl2-supplemented native PAGE analysis of 210-bp-DNA template assemblies. Lane 1: linear ssDNA strand; lane 2: circularized ssDNA strand; lane 3: dsDNA [21+]20-circle; lane 4: dsDNA [21−]20-circle.