Abstract
Telomeric repeat binding factor 2 (TRF2) folds human telomeres into loops to prevent unwanted DNA repair and chromosome end-joining. The N-terminal basic domain of TRF2 (B-domain) protects the telomeric displacement loop (D-loop) from cleavage by endonucleases. Repressor activator protein 1 (Rap1) binds TRF2 and improves telomeric DNA recognition. We found that the B-domain of TRF2 stabilized the D-loop and thus reduced unwinding by BLM and RPA, whereas the formation of the Rap1–TRF2 complex restored DNA unwinding. To understand how the B-domain of TRF2 affects DNA binding and D-loop processing, we analyzed DNA binding of full-length TRF2 and a truncated TRF2 construct lacking the B-domain. We quantified how the B-domain improves TRF2’s interaction with DNA via enhanced long-range electrostatic interactions. We developed a structural envelope model of the B-domain bound on DNA. The model revealed that the B-domain is flexible in solution but becomes rigid upon binding to telomeric DNA. We proposed a mechanism for how the B-domain stabilizes the D-loop.
INTRODUCTION
Telomeres are nucleoprotein structures that protect chromosomal ends from undesirable nucleolytic degradation, recombination, DNA repair processes and lethal chromosome fusions. Telomeric DNA repeats are maintained by telomerase that adds repetitive DNA sequences to the very end of a chromosome (1). The telomere maintenance prevents gene loss by addressing two major challenges of cell survival: the incomplete replication of DNA 3′strand after each cell division and the unwanted recognition of naked DNA chromosome tips as double strand breaks. Telomeric DNA forms loops that hide and protect the very ends of chromosomal DNA. In such telomere loop (t-loop), the double-stranded DNA twists around forming a lasso-like structure (2). In addition, the single-stranded 3′terminus inserts back inside the double-stranded DNA and forms a displacement loop (D-loop). As a result, telomere loops hide chromosome termini from unrestricted protein recognition of telomere ends as double-strand breaks (3). However, the loop formation has to be reversible as telomere loops must open when DNA extension or replication take place. The exact mechanism how the D-loop opens during telomere replication remains elusive.
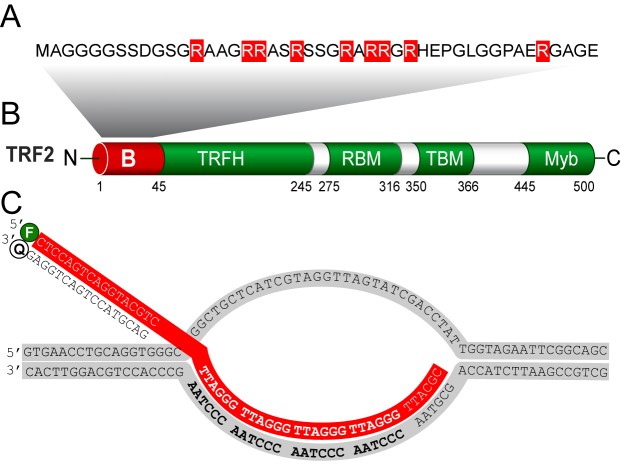
What we know is the mechanism how a specific six-protein complex called shelterin mediates protection of telomeres. Shelterin covers telomeres and regulates access of telomerase and other proteins to telomeric DNA in human germ and stem cells (4). Shelterin proteins are well evolutionary conserved as they regulate telomerase recruitment in mammals and in yeasts as well (5). Telomeric repeat binding factor 2 (TRF2) plays a central role in the loop formation and mediates different stages of telomere protection (6). Regarding the structure, human TRF2 comprises an N-terminal basic domain (B-domain) that is rich in glycine and positive arginine residues, a TRF homology (TRFH) domain that mediates homodimerization, a flexible linker containing Rap1 binding motif (RCT), and a C-terminal Myb domain that selectively recognizes 5′YTAGGGTTR double-strand DNA sequence (Figure 1B) (7).
Figure 1.
Structures of the B-domain, TRF2 and D-loop. (A) The B-domain comprises nine positively charged arginine residues (highlighted) that are evolutionarily conserved between mouse and human (39). (B) The B-domain is located at the N-terminus of TRF2. Additionally, TRF2 consists of TRFH dimerization domain, Rap1-binding motif RBM, TIN2-binding motif TBM and C-terminal DNA binding Myb domain that selectively recognizes telomeric DNA. (C) The sequence of the D-loop contains four telomeric repeats in the invading strand. The invading strand was labeled with fluorescein (F). The complementary strand with quencher OQA (Q) was added exclusively for thermal stability measurements.
Regarding the biological functions, TRF2 mediates t-loop formation (2) and protects telomeric Holliday junctions (8). Additionally, TRF2 might mediate telomeric DNA wrapping and thus compacting of telomeric chromatin (9). Recent studies showed that the B-domain is indispensable for essential protective functions of TRF2 (10). In human cells, the Gilson laboratory showed that the B-domain of TRF2 prevents MUS81, GEN1 and SLX4-associated endonucleases to process telomere replication intermediates such as loops or stalled forks (11). The studies in humanized yeasts suggest that the evolutionally conserved B-domain (Figure 1A) regulates the resolvase activities in order to repair stalled forks occurring during telomere replication (11). Human DNA helicases such as the Bloom syndrome helicase (BLM) on the first place and also WRN, RecQL4, RecQL1, RTEL1 have been tightly implicated in telomere maintenance mediated by TRF2 (12,13). BLM preferentially processes telomeric DNA intermediates in the presence of SLX1-SLX4 nuclease, as has been shown in vivo and in vitro (14). The BLM preference for D-loop processing suggests that BLM plays a master role in dissolving telomeric loops. Similarly, Replication Protein A (RPA) also participates in D-loop processing. RPA is a heterotrimeric complex that binds to single-stranded DNA and is essential for initiation and elongation of DNA replication. RPA can unwind dsDNA for bona fide DNA helicase activity that might take part in the initiation of DNA replication in eukaryotes (15).
Recently, we showed that the complexation of the two shelterin proteins TRF2 and Rap1 improves TRF2 binding specificity toward telomeric DNA (16). Human Rap1 is the closest binding partner of TRF2. Rap1 mediates genome stability as it inhibits non-homologous end-joining at telomeres (17). Our quantitative biology experiments and available structural data suggest that Rap1 prevents non-specific DNA binding of TRF2 via its B-domain. Even though cytological studies have proven the significance of the B-domain of TRF2 for loop disassembling (13) and chromatin compacting (18), there are only limited structural and quantitative studies that describe thoroughly how the B-domain of TRF2 affects DNA loop binding and processing. The main hypothesis is that the B-domain of TRF2 could stabilize D-loops and thus the B-domain lessens unwinding of chromosome ends. Additionally, we wonder whether Rap1 could affect the engagement of the TRF2 B-domain in D-loop stabilization and DNA processing.
In this study, we addressed the following questions. How does the TRF2 B-domain affect unwinding of telomeric and non-telomeric D-loops? What is the role of Rap1 in D-loop processing? What is the mechanism for how the B-domain contributes to overall binding affinity of TRF2 to telomeric DNA?
Here we show that the B-domain of TRF2 stabilizes the telomeric D-loop and thus reduces unwinding of dsDNA by BLM and RPA. Further, we reveal that human shelterin protein Rap1 upon complexation with TRF2 restores D-loop susceptibility to unwinding. Additionally, we show that the B-domain doubles the binding affinity of TRF2 for telomeric DNA and the affinity increase is due to mainly electrostatic forces. Further, we demonstrate that the flexible B-domain becomes rigid and structured when bound to DNA, which explains the increased D-loop stability and reduced unwinding in the presence of TRF2. Finally, we propose a model of domain arrangement of the shelterin proteins TRF2 and Rap1 that suggests how the B-domain of TRF2 stabilizes D-loops and similar DNA structures.
MATERIALS AND METHODS
Cloning, expression and purification of TRF2 variants
The cDNA sequence of TRF2 was synthesized by Source BioScience and used for construction of full-length TRF2 and the N-terminal B-domain of TRF2 (B-domain). The mutant version of TRF2 lacking the B-domain (ΔBTRF2) was prepared from a pcDNA N-Myc plasmid containing ΔBTRF2 gene (kindly provided by Titia de Lange, The Rockefeller University, NY, USA). Trf2 and ΔBtrf2 were cloned to pDONR/Zeo vector (Life Technologies) using two sets of primers (Table 1) and BP clonase enzyme mix from Gateway technology (Life Technologies). The resulting plasmids pDONR/Zeotrf2/ΔBtrf2 were cloned into the pHGWA expression vector (19) in a recombination reaction using LR clonase enzyme mix (Life Technologies) and expressed as His-tagged proteins in Escherichia coli BL21(DE3) (TRF2) and BL21(DE3)RIPL (ΔBTRF2). The B-domain nucleotide sequence was cloned into BamHI and XhoI sites of pET28bSMT3 vector using primers shown in Table 1 and expressed in E. coli BL21(DE3)RIPL.
Table 1. Sequences of primers used for the construction and cloning of TRF2 variants.
| Cloned sequence | Primer | Sequence of primer 5′-3′ |
|---|---|---|
| TRF2 | Forward 1 | CTGGAAGTTCTGTTCCAGGGGCCCATGGCGGGAGGAGGCGGGAGTAGC |
| TRF2/ΔBTRF2 | Forward 2 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCCTGGAAGTTCTGTTCCAGGGGCCC |
| TRF2 | Reverse | GGGGACCACTTTGTACAAGAAAGCTGGGTTCATCAGTTCATGCCAAGTCTTTTCAGTGT |
| ΔBTRF2 | Forward 1 | CTGGAAGTTCTGTTCCAGGGGCCCGAGGCACGGCTAAGAGGCAGTC |
| ΔBTRF2 | Reverse | GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAGTTCATGCCAAGTCTTTTCATGGT |
| B-domain | Forward | ATAGGATCCATGGCGGGAGGAGGCGG |
| B-domain | Reverse | ATATATCTCGAGCTACCCCGCGCCGCGCTC |
BL21(DE3) cells harboring the pHGWAtrf2 plasmid were grown in Luria-Bertani medium containing 100 μg ml−1 ampicillin at 37°C until A600 reached 1.0. The cells were cultured for 3 h at 25°C after the addition of IPTG (isopropyl-β-D-thiogalactopyranoside) (Molekula) to a final concentration of 1 mM. Escherichia coli BL21(DE3)RIPL with pHGWAtrf2ΔBwere grown in Terrific Broth medium containing 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol at 37°C, [160 rpm] until A600 reached 0.5. The protein expression was initiated by the addition of IPTG to a final concentration of 0.5 mM, and the cells were incubated at 22°C for the next 5 h.
Cells carrying plasmid with the B-domain insert were grown in minimal medium (40 mM Na2HPO4·2H2O, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 2 mM MgSO4, 0,1 mM CaCl2, 0,4% glucose). The expression medium contained 50 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol. Cells were cultivated at 37°C, [160 rpm] until A600 reached 0.5. After reducing the temperature to 25°C and addition of IPTG to 0.5 mM, the cells were incubated for next 5 h and harvested by centrifugation at 8000 g for 8 min at 4°C. The pellet was dissolved in buffer containing 50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 10 mM imidazole, 0.5% Tween-20, 10% glycerol, 5 μM Leupeptin, 4 μM Pepstatin A (Applichem) along with protease inhibitor cocktail cOmplete tablets EDTA-free (Roche). The cell suspension was sonicated for 3 min of process time with 1 s pulse and 2 s of cooling on ice (Misonix). Cell supernatant was collected after centrifugation at 20 000 g, 4°C for 1 h.
Recombinant TRF2 variants were purified by immobilized-metal affinity chromatography using TALON®metal affinity resin (Clontech). Filtered protein extracts (0.45 μm filter) were loaded on a column pre-equilibrated by 50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 10 mM imidazole, 0.5% Tween-20, 10% glycerol buffer. Proteins of our interest were eluted at 300 mM imidazole in the same buffer without Tween-20. The B-domain eluate was incubated overnight at 4°C with Ulp1 protease to remove tags during dialysis against a buffer containing 50 mM sodium phosphate pH 7.0 and 200 mM NaCl. After cleavage, the sample was loaded onto TALON®metal affinity resin (Clontech). Unbound fractions were loaded onto the HiLoad 16/600 column containing Superdex75 pg (GE Healthcare Life Sciences) and resolved using 50 mM sodium phosphate buffer pH 7.0 with 200 mM NaCl.
The proteins were concentrated and the buffer was exchanged to 50 mM sodium phosphate pH 7.0 and 50 mM NaCl by ultrafiltration (Amicon 3K/30K, Millipore). The concentration of purified proteins was determined by Bradford assay. We evaluated protein purity by SDS-polyacrylamide gels stained by Bio-Safe Coomassie G 250 (Bio-Rad). Western blotting, using monoclonal Anti-polyHistidine antibody produced in mouse and Fc-specific Anti-Mouse IgG—peroxidase antibody produced in goat (Sigma Aldrich), confirmed the presence of the tagged proteins of our interest. Quantitative mass spectrometry analyses confirmed appropriate purity of the obtained proteins for further studies.
DNA substrates
For the D-loop unwinding assay, a synthetic telomeric D-loop substrate (D-loop R4) was prepared by mixing an equimolar amount of the oligonucleotides 1 and 2 (Table 2) in the hybridization buffer 25 mM Tris–HCl pH 7.5, 50 mM LiCl (to avoid quadruplex formations), 10 mM MgCl2, heated at 90°C for 3 min and cooled to room temperature. Then, to make the 3′ ssDNA invasion an equimolar amount of 5′ FITC-labeled oligonucleotide 3 was added. The mixture was incubated at 37°C for 20 min and cooled slowly (∼1°C/min) to room temperature to allow DNA annealing. The substrate was purified using a Mono Q 5/50 GL column (GE Healthcare) with a gradient of 50–1000 mM LiCl in 25 mM Tris–HCl, pH 7.5. D-loop fractions were dialyzed into 25 mM Tris–HCl, pH 7.5, 3 mM MgCl2 and concentrated in an Amicon 3K (Millipore). The non-telomeric DNA substrate (D-loop N) was prepared exactly as described by Youds et al. (20).
Table 2. Oligonucleotides used for the construction of telomeric D-loop R4.
| Sequence of primer 5′-3′ | Modification | |
|---|---|---|
| Oligonucleotide 1 | GTGAACCTGCAGGTGGGCGGCTGCTCATCGTAGGTTAGTATCGACCTATTGGTAGAATTCGGCAGC | |
| Oligonucleotide 2 | GCTGCCGAATTCTACCAGCGTTAACCCTAACCCTAACCCTAACCCTAAGCCCACCTGCAGGTTCAC | |
| Oligonucleotide 3 | CTCCAGTCAGGTACGTCTTAGGGTTAGGGTTAGGGTTAGGGTTACGC | 5′ FITC |
| Oligonucleotide 4 | GACGTACCTGACTGGAG | 3′ OQA |
For the DNA binding affinity studies, two DNA duplexes R2 and R5 were prepared by annealing a fluorescently labeled oligonucleotide (Alexa Fluor 488) with the sequences 5′-GTTAGGGTTAGGGTTAG-3′ and 5′-GTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAG-3′, respectively and their complementary strand. The sequence of R2 was designed in accordance to the optimal binding site of TRF2 defined by the de Lange laboratory (21). The incubation time and buffers were the same as described above. All oligonucleotides were purchased from Sigma-Aldrich.
Fluorescence anisotropy
Measurements of TRF2 variants binding to telomeric DNA by fluorescence anisotropy were performed on a FluoroMax-4 spectrofluorometer (Horiba Jobin Yvon, Edison, NJ, USA) equipped with a temperature controlled cuvette holder. All experiments were carried out at 25°C in a quartz-glass cuvette with optical length of 10 mm. Fluorescence anisotropy of oligonucleotides labeled with Alexa Fluor 488 was monitored at an excitation wavelength of 493 nm and emission wavelength of 516 nm. The slit width (both excitation and emission) for all measurements was 9 nm and the integration time was 1 s. The cuvette contained 1.4 ml of DNA duplex R2 or R5 (7.5 nM) in a buffer containing 50 mM sodium phosphate pH 7.0 and 50 mM NaCl if not stated otherwise. A protein aliquot was titrated into the DNA solution in the cuvette and measured after a 2 min incubation. Fluorescence anisotropy was measured three times and averaged with relative standard deviation always lower than 3%. The values of dissociation constants were determined by non-linear least square fits according to the equation r = rMAX c/(Kd+c) using SigmaPlot 12 (Systat Software) and confirmed by symbolic equation-based fitting using Dynafit (22).
Electrostatic component of binding
To reveal the contribution of the electrostatic component to the binding of TRF2 to telomeric DNA, we measured the binding of TRF2 and ΔBTRF2 to DNA at different concentrations of NaCl. The electrostatic component of binding comes from the formation of ion pairs between the positive charge of amino acid residues and the negative charge of DNA. The electrostatic component of binding was determined from the linear dependence of the binding constant on ionic strength, as described in our previous studies (16,23).
SAXS analysis and shape reconstitution
The SAXS (Small-Angle X-ray Scattering) was measured on the BioSAXS-1000, Rigaku at CEITEC (Brno, Czech Republic). SAXS data were collected at 277.15 K with X-ray beam wavelength 1.54 Å. The distance between the sample and the detector (PILATUS 100K, Dectris Ltd.) was 0.48 m covering a scattering vector (q = 4πsin(θ)/λ) range from 0.009 to 0.65 Å-1. For solvent and sample, one two-dimensional image was collected with 1-h exposure time per image. Radial averaging of two-dimensional scattering images and the solvent subtractions were performed using SAXSLab3.0.0r1, Rigaku. All datasets were truncated to maximum scattering vector of 0.3 Å-1 for further analysis. Scattering-derived parameters were determined using PRIMUS from ATSAS v2.6.0. The evaluation of the theoretical solution scattering of the crystal structure of the telomeric duplex (PDB ID: 1W0T) and fitting to experimental data was performed by CRYSOL v2.8.3, where the automatic constant subtraction was allowed, while other parameters were kept default. The data from the telomeric DNA duplex and data of the DNA–peptide complex were used for the two-phase ab initio shape reconstruction performed by MONSA v1.45, where the search sphere diameter was set to 40 Å, volumes of individual phases were set to 13 520 Å3 for DNA and 4310 Å3 for protein, regarding the Porod volumes. The superimposition of the ab initio SAXS envelopes and the atomic models was performed by SUPCOMB v23. The graphical representation of ab initio SAXS models was generated using UCSF Chimera v1.10.1.
D-loop unwinding assay
The assay has been performed according to helicase reaction protocols published previously (20,24). D-loop substrates (3 nM) fluorescently labeled with fluorescein (FITC) were incubated with TRF2, ΔBTRF2 or Rap1–TRF2 (750 nM) at 24°C for 5 min in the reaction buffer (25 mM Tris-acetate, pH 7.5, 5 mM CaCl2, 2 mM MgCl2, 5 mM ATP, 5 mM DTT and 100 μg/ml BSA) containing an ATP regenerating system consisting of 20 mM creatine phosphate and 20 μg/ml creatine kinase. The human ssDNA binding factor RPA was prepared as described by Sigurdsson et al. (25). RPA was added to final concentrations as indicated in figure legends and incubated for 5 min at 24°C. Finally, BLM (160 nM) was added and the mixture of total volume 10 μl was incubated at 37°C for 20 min. BLM (fragment 642–1290 aa) was a generous gift of Victoria Marini and Mate Gyimesi prepared according to Janscak et al. (26).
The reaction was stopped by the addition of 1 μl of deproitenization buffer (final concentration: 0.3% SDS and 1 mg/ml proteinase K), followed by a 5 min incubation at 37°C. After deproteinization, the reactions were mixed with 2 μl of 6× loading buffer (60% glycerol; 10 mM Tris–HCl, pH 7.6 and 60 mM EDTA). In the RPA titration experiment, RPA concentrations varied in the range 0–1400 nM. The incubation time and buffers were the same as described above. For all assay arrangements, DNA products were analyzed by electrophoresis on a vertical 10% native polyacrylamide gel in 1× TBE buffer. The electrophoresis proceeded at 9.5 V/cm for 80 min at 4°C. The gels were visualized in a fluorescent scanner Typhoon FLA 9500 and quantified by ImageQuant TL software (version 8.1; GE Healthcare).
Thermal stabilization measurement
The thermal stability of D-loop structures were measured by a FluoroMax-4 spectrofluorometer (Horiba Jobin Yvon, Edison, NJ, USA) equipped with a water bath controlling the temperature of the sample chamber. All experiments were carried out in the temperature range from 10 to 90°C in a quartz-glass cuvette with optical length of 10 mm. Fluorescence intensity of D-loop R4 (FITC labeled) was monitored at excitation wavelength 493 nm and emission wavelength 516 nm. Excitation and emission slits were 5 nm. The integration time was 0.5 s. The cuvette contained 1000 μl of labeled D-loop R4 (4 nM) with the hybridized complementary invading strand (quencher OQA label) (see Figure 1C and Table 2) in 50 mM sodium phosphate pH 7.0, 50 mM NaCl in the absence or presence of TRF2/ΔBTRF2/ΔMybTRF2 at 500 nM concentration to ensure that all possible binding sites on the D-loop are occupied. The dissociation of FITC invading strand from the quenching strand was observed as a fluorescence increase, which corresponds to the ratio of free and hybridized (quenched) invading strand of D-loop R4. Melting curves were normalized to the fluorescence intensity at 25°C. The average heating gradient was 2°C/min. The melting temperature (Tm) was determined as the temperature corresponding to the maximum of the derivation dFI/dT.
RESULTS
The B-domain of TRF2 reduces D-loop unwinding
To describe how the TRF2 B-domain affects D-loop unwinding, we quantified the unwinding by BLM helicase in the presence of either full-length TRF2 or ΔBTRF2. As a DNA substrate, we used two types of D-loop constructs. The first was D-loop R4 containing four full telomeric repeats in the inner duplex part of the bubble as shown in Figure 1C. The sequence of D-loop R4 was based on reported studies from the Bohr laboratory (24,27). The non-telomeric D-loop N was the same as designed by the Boulton laboratory (20). In all experiments with TRF2 and BLM, we used optimized RPA concentration sufficient to prevent re-hybridization of unwound single-strands and to minimize the natural unwinding by RPA (Figure 2A and C, lane 7). We verified that D-loop substrates are stable at 3 nM without any sign of unwinding in the presence of RPA at 175 nM concentration and lower (Supplementary Figure S1). Thus, we confirmed that RPA has no effect on D-loop unwinding at 160 nM concentration as used for BLM unwinding experiments.
Figure 2.
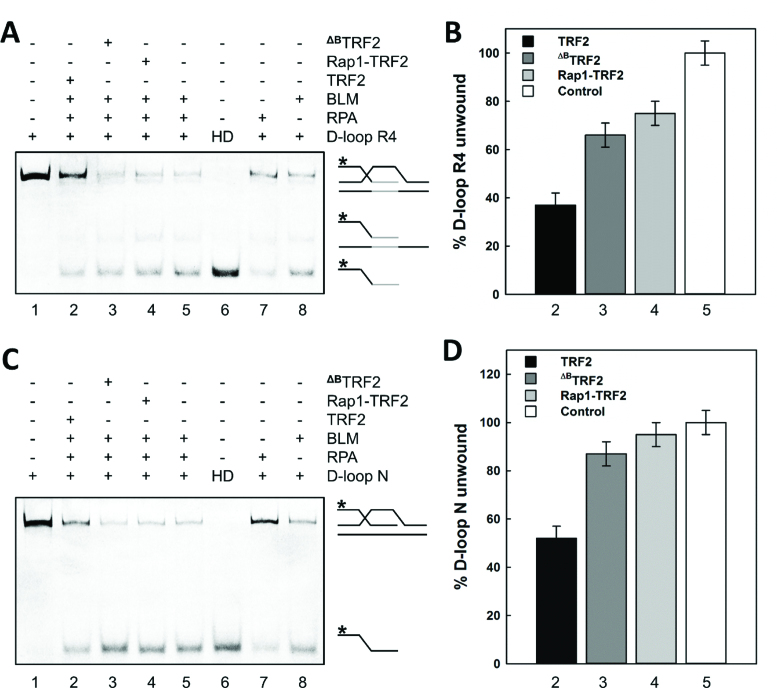
The B-domain of TRF2 reduces unwinding of D-loops by BLM whereas Rap1 restores DNA unwinding. (A) D-loop R4 containing four telomeric repeats or (C) non-telomeric D-loop N was unwound by helicase BLM. Reaction mixtures (10 μl) contained D-loop (3 nM) labeled by fluorescein and incubated with BLM (180 nM) in the presence of RPA (160 nM; lane 5) and either TRF2, ΔBTRF2 or Rap1–TRF2 complex (750 nM; lanes 2,3,4). D-loop only (lane 1); HD heat denatured D-loop (lane 6); D-loop incubated either with RPA only (lane 7) or BLM only (lane 8) were used as control samples. An asterisk denotes the fluorescently labeled invasion strand of the D-loop. Telomeric repeats are shown in gray. Products of D-loop unwinding assay were analyzed on a vertical 10% native polyacrylamide gel. (B) The bar graph exhibits the percentage of D-loop R4 unwinding of selected reactions from part A. The lane 5 was assumed as 100% unwinding which corresponds to the open column denominated as Control. (D) The bar graph demonstrates the percentage of D-loop N unwinding of selected reactions from part C. The error bars represent the mean and standard deviation from three independent experiments.
We observed that TRF2 pre-bound to D-loop R4 reduced DNA unwinding of BLM to 37% of the original level (compare lanes 5 and 2 in Figure 2B). Similarly, TRF2 on D-loop N reduced DNA unwinding of BLM to 52% when compared to naked D-loop N (compare lanes 5 and 2 in Figure 2D). Upon pre-incubation of ΔBTRF2 with D-loops, we observed that the extent of BLM unwinding decreased only to 66% for D-loop R4 and to 87% for D-loop N (compare lanes 5 and 3 in Figure 2B and D). In other words, the extent of unwinding was restored close to the original level when the B-domain was removed from TRF2.
To address whether the B-domain induced reduction in D-loop unwinding is related to the BLM–TRF2 interaction (24), we tested the effect of TRF2 on D-loop unwinding in the absence of BLM. As RPA has been shown to unwind dsDNA (28), we assessed the effect of TRF2 on the unwinding of D-loops by RPA without the presence of any other helicase. After the pre-incubation of full-length TRF2 with D-loops and upon subsequent RPA addition, we observed that TRF2 significantly reduced the natural unwinding activity of RPA. Specifically, we observed that full length TRF2 on D-loop R4 and D-loop N decreased DNA unwinding to 34 and 7%, respectively (compare lanes 5 and 2 in Supplementary Figure S2). Notably, in the absence of the B-domain, pre-incubation of ΔBTRF2 with D-loops decreased RPA unwound fractions only to 61% for D-loop R4 and to 23% for D-loop N (compare lane 5, 3 in Supplementary Figure S2). In summary, RPA induced D-loop unwinding was significantly more pronounced in the absence of the B-domain of TRF2 than in its presence.
Altogether, TRF2 prevented D-loop unwinding by BLM or RPA. Our unwinding experiments further suggested that the B-domain of TRF2 contributed to tightening of D-loop structures regardless of whether BLM helicase or general single-strand DNA binding protein RPA with helicase activity was allowed to unwind D-loops.
Rap1 in complex with TRF2 restores full D-loop unwinding
Additionally, we tested how the closest TRF2 binding partner Rap1 affects D-loop unwinding. We observed that in the presence of Rap1–TRF2 complex, BLM unwinding of D-loop R4 or D-loop N was 97 and 83%, respectively. Thus, Rap1 complexed with full-length TRF2 affected BLM unwinding analogically to the extent that we observed when the B-domain was removed from TRF2 (compare lanes 3 and 4 in Figure 2B and D). Similarly, upon pre-incubation of Rap1–TRF2 complex with D-loop R4 and N, RPA unwinding levels came back to 60 and 22%, respectively; i.e. similar unwinding levels as observed for ΔBTRF2 (compare lanes 3 and 4 in Supplementary Figure S2). In other words, Rap1–TRF2 complex restored D-loop unwinding of RPA and BLM close to the extent as if there were no TRF2 bound to DNA. Thus, similar unwinding propensity for Rap1–TRF2 and ΔBTRF2 suggests that Rap1 prevented the inhibition of unwinding mediated by the B-domain of TRF2.
The B-domain of TRF2 is essential for telomeric D-loop stabilization
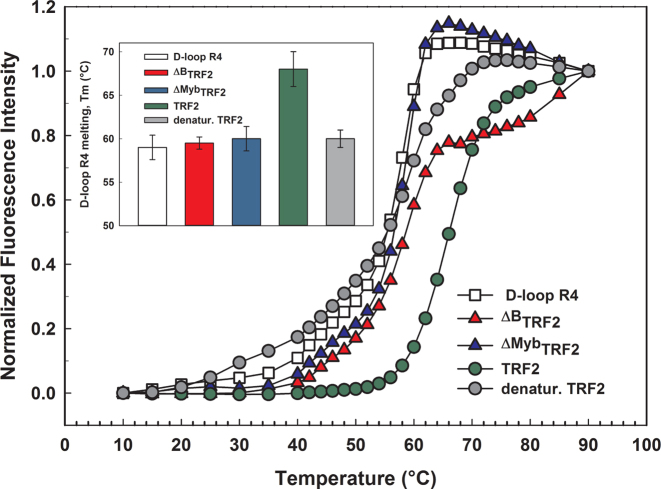
To reveal the mechanism of how TRF2 prevents D-loop unwinding, we quantified how TRF2 affects thermal stability of the telomeric D-loop R4. Before our D-loop melting studies, we measured thermal stability of all used TRF2 variants by differential scanning fluorimetry. The denaturation temperature for all the TRF2 variants was above 52°C (thermal data not shown). To monitor strand dissociation during unwinding, we hybridized the FITC labeled invading strand with the quencher attached complementary strand (Figure 1C and Table 2). We used fluorescence spectroscopy to detect the temperature-induced dissociation of the FITC labeled invading strand from the complementary quenching strand. Thus, the fluorescence intensity of FITC was proportional to the ratio of free and hybridized invading strand of D-loop R4. To determine the direct contribution of the B-domain on the thermal stability of D-loop R4, we allowed TRF2 or ΔBTRF2 to bind D-loop R4. We carried out equilibrium melting of D-loop R4 in the presence or absence of the TRF2 variants. The resulting melting curves were differentiated to obtain well-defined melting temperatures. The melting temperature was determined as the temperature at which the derivation reaches maximum.
We observed that full-length TRF2 bound to D-loop R4 elevated the thermal stability by 8°C. The B-domain removal in case of ΔBTRF2 caused the increase of thermal stability only by 2°C (Figure 3). The thermal stability increase observed for ΔBTRF2 is close to the edge of confidence interval of melting temperature determination. The variant lacking the Myb domain ΔMybTRF2, and denatured TRF2 showed no effects on D-loop thermal stability. Similarly, we observed no significant effect of Rap1–TRF2 complex on D-loop thermal stability (data not shown). Altogether, our quantitative fluorescence measurements of thermal stability suggested that mainly the B-domain of TRF2 stabilizes the telomeric D-loop.
Figure 3.
TRF2 stabilizes the D-loop via its B-domain. Equilibrium thermal melting curves for D-loop R4 (open square), and D-loop R4 in the presence of either TRF2 (upper circle in legend), ΔBTRF2 (upper triangle in legend), heat-denatured TRF2 (lower circle in legend) or ΔMybTRF2 (lower triangle in legend) in 50 mM sodium phosphate buffer pH 7, 50 mM NaCl. The closed-to-open transition of the invading strand was monitored by the fluorescence intensity of FITC that increased when the complementary quencher labeled strand dissociated. The inset with the bar graph shows the melting temperatures determined from the first derivations of melting curves.
The B-domain increases two-fold the DNA binding affinity of TRF2
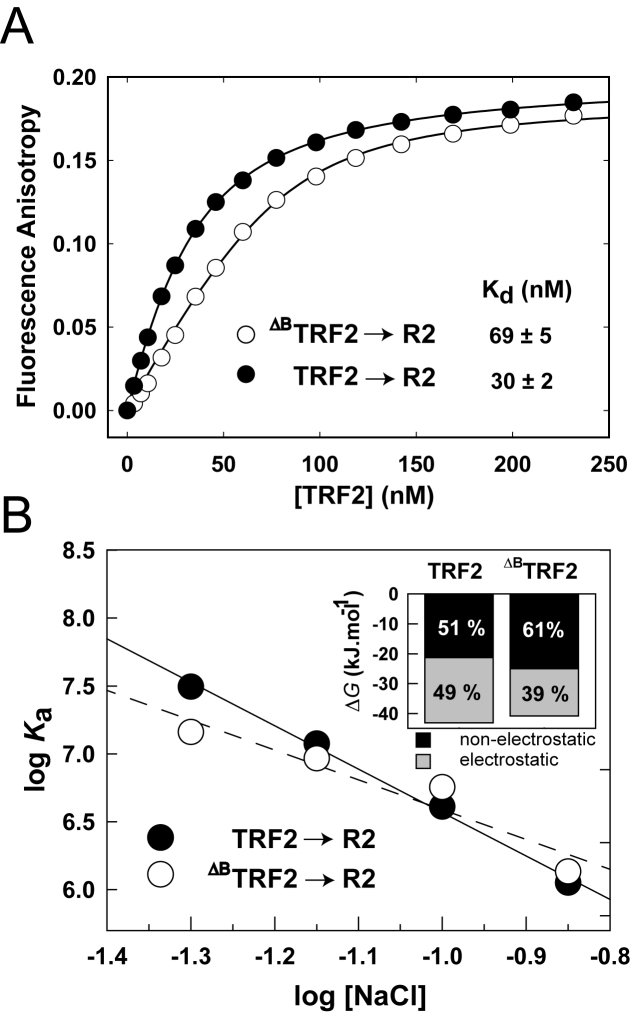
To quantify how the N-terminal domain of TRF2 contribute to the binding affinity of whole TRF2 to telomeric DNA, we measured equilibrium binding curves of full-length TRF2 and ΔBTRF2 to DNA by fluorescence anisotropy. To describe the B-domain binding effect thoroughly, we used telomeric DNA duplexes of different lengths containing either two complete telomeric repeats (R2) or five telomeric repeats (R5). The DNA binding isotherms obtained for full-length TRF2 and ΔBTRF2 are shown in Figure 4A and Supplementary Figure S4. We observed a significant increase of the binding affinity of full-length TRF2 for telomeric DNA when compared to DNA binding affinity of ΔBTRF2. The comparison of the equilibrium dissociation constants revealed two-fold lower Kd for full-length TRF2 when compared to Kd of the truncated variant ΔBTRF2. The binding experiments were performed in 50 mM sodium phosphate pH 7.0 containing 50 mM NaCl.
Figure 4.
The B-domain increases TRF2 binding affinity to telomeric DNA via increased electrostatic attraction. (A) TRF2 or ΔBTRF2 (5 μM) was titrated to Alexa Fluor 488 labeled DNA duplex (7.5 nM) in 50 mM sodium phosphate, pH 7.0, 50 mM NaCl at 25°C. Binding isotherms of full-length TRF2 (closed circles) and truncated ΔBTRF2 (open circles) binding to telomeric DNA duplex R2 containing two telomeric repeats were measured by fluorescence anisotropy. (B) The B-domain of TRF2 enlarges the electrostatic attraction to telomeric DNA. The dependence of logarithm of the association constant for binding of TRF2 or ΔBTRF2 to telomeric DNA duplex (7.5 nM) on logarithms of NaCl concentration. NaCl concentration ranged from 50 to 140 mM in 50 mM sodium phosphate buffer pH 7.0. The inset bar graph shows the relative contribution of electrostatic and non-electrostatic interactions to the total free energy of binding of TRF2 or ΔBTRF2 to telomeric DNA.
To confirm that this buffer is fully compatible with the physiologically relevant buffer containing 110 mM NaCl and 10 mM sodium phosphate, pH 7.0 (Supplementary Figure S3), we recorded binding curves in both buffers. The binding curve analysis showed that TRF2 binds telomeric DNA in 50 mM sodium phosphate pH 7.0 containing 50 mM NaCl with the same binding affinity as in 10 mM sodium phosphate, pH 7.0 with 110 mM NaCl. The comparison of Kd values revealed that DNA binding affinities measured in two buffers was comparable and confirmed previously measured DNA binding difference between TRF2 and ΔBTRF2. Thus, our binding affinity measurements suggested that the B-domain doubles the overall TRF2 binding affinity to DNA in physiologically relevant conditions.
The B-domain enhances non-specific electrostatic attraction of TRF2 to telomeric DNA
To reveal the origin of the B-domain contribution to DNA binding affinity of TRF2, we analyzed the electrostatic component of TRF2–DNA interactions. We measured binding of full-length TRF2 and ΔBTRF2 to telomeric DNA duplex R2 at NaCl concentrations ranging from 50 to 140 mM in 50 mM sodium phosphate buffer pH 7.0. The logarithmic dependence of the equilibrium binding constants of TRF2 and ΔBTRF2 on logarithm of salt concentration is shown in Figure 4B. From the slope of the linear regression, we determined the Z parameter (Supplementary Table S1). The Z parameter corresponds to the number of ionic bonds between the protein and DNA formed upon binding. The binding of TRF2 or ΔBTRF2 to telomeric DNA duplex R2 induces formation of 4.6 and 3.4 ion pairs on the average, respectively. The higher number of ionic pairs formed between full-length TRF2 and DNA when compared with ΔBTRF2-DNA binding means the higher electrostatic attraction to telomeric DNA caused by the B-domain.
Based on the salt dependencies of binding affinities, we determined how the B-domain contributes to the total thermodynamic stability of TRF2 on DNA. We calculated the value of the non-electrostatic contribution of binding log Kanel from the extrapolated y-intercept of the linear dependence for both full length and truncated TRF2. The electrostatic contribution of binding log Kael was calculated after subtraction of non-electrostatic log Kanel from total log Ka as shown previously (16). When the B-domain was removed from TRF2, the electrostatic contribution to the DNA binding affinity dropped by 5.8 kJ/mol. The decrease of electrostatic attraction was partly compensated by an increase of non-electrostatic contribution to binding. In total, we have found that the B-domain contributes to DNA binding by 2.1 kJ/mol from total free energy 42.9 kJ/mol (Supplementary Table S2).
Additionally, we calculated how the B-domain changed the ratio of the electrostatic and non-electrostatic contributions to DNA binding of TRF2. In general, the electrostatic component of binding is associated with nonspecific attraction of opposite charges on the protein and DNA; the non-electrostatic component of binding is associated with the formation of specific bonds between the protein and DNA. The electrostatic and non-electrostatic components of the binding energy for TRF2 and ΔBTRF2 to DNA are shown in the inset of Figure 4B. The B-domain interactions contribute to the increase of electrostatic part of the total energy of binding from 39% for ΔBTRF2 to 49% for full-length TRF2. We conclude that the B-domain was responsible for one fifth of the overall electrostatic contribution to the total free energy of TRF2 binding to telomeric DNA.
The flexible basic domain of TRF2 becomes rigid upon binding to telomeric DNA duplex
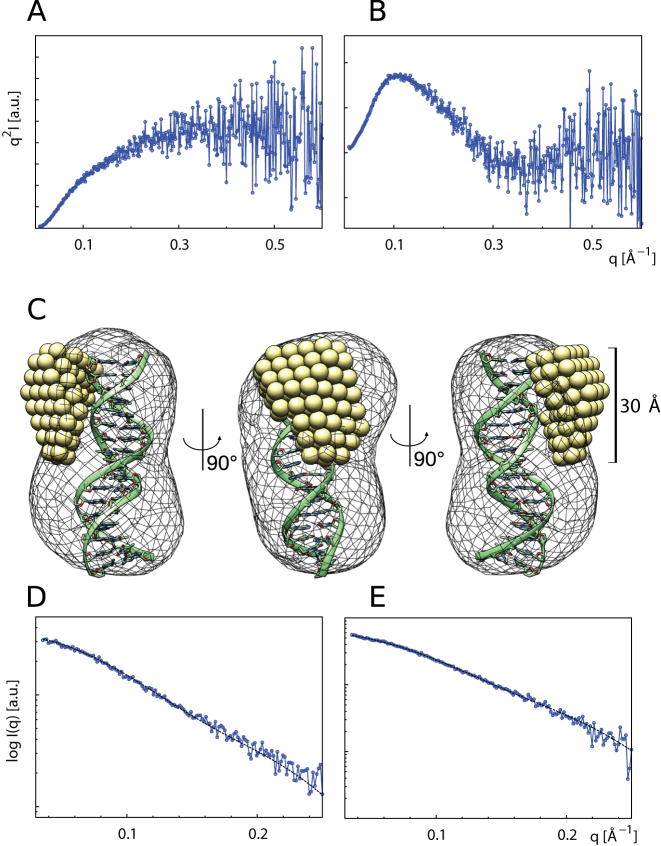
To address the structural arrangement of the B-domain when interacting with telomeric DNA duplex, we carried out SAXS measurements with the isolated B-domain and DNA duplex R2. The SAXS data were collected for the telomeric DNA duplex R2, B-domain and B-domain-DNA complex at 277K (Figure 5A and B). To prevent a quadruplex formation, LiCl to final 50 mM was added into 20 mM Tris–HCl buffer pH 7.5. SAXS derived parameters are summarized in Supplementary Table S3. The hydrated volume of the particle (Porod volume) was determined as 4310 Å3 for the B-domain, 13 520 Å3 for the DNA duplex and 18530 Å3 for the B-domain–DNA complex. The expectedly increased Porod volume suggested that the B-domain formed a stable complex with the DNA duplex in solution.
Figure 5.
The B-domain of TRF2 becomes rigid upon binding to telomeric DNA duplex. Kratky plots (I*q2 versus q) of the unbound B-domain (A) and B-domain in complex with telomeric DNA duplex (B). Scattering data from the unbound B-domain (A) exhibit increase characteristic for macromolecules with substantial flexible or unstructured regions, while scattering data from the B-domain–DNA complex (B) display clear minimum characteristic for well-folded compact particles. (C) The typical MONSA model of the B-domain–DNA complex. The DNA phase (mesh) is superimposed with molecular model of telomeric DNA duplex. The protein phase (spheres) illustrates the bound B-domain as a compact body. Fits of simulated scattering to experimental data: (D) DNA phase with final χ2 value 1.22. (E) B-domain–DNA complex phase with χ2 value 1.06.
Additionally, we evaluated the theoretical solution scattering of the crystal structure of the telomeric DNA duplex. The molecular model of the telomeric duplex was adapted from PDB entry id: 1W0T (7), where two initial nucleotides were removed from the original DNA duplex. The overall CRYSOL model fits well with the χ2 value 1.02 (data not shown). Computed structural parameters of the molecular model as envelope Rg of ∼18Å, envelope volume 13 740 Å3 and envelope diameter ∼64 Å are well comparable with basic SAXS parameters derived from experimental data.
As Kratky plots indicate, the free B-domain was flexible in solution. However, if the B-domain bound DNA, the flexible nature of the peptide chain was lost and the B-domain became rigid. The loss of flexibility is further supported by comparison of the maximum distance values ∼78 Å of the free B-domain versus ∼58 Å of the B-domain complexed with DNA. The IUPred on-line service (29) was used to predict disordered regions of the peptide. All 44 amino-acid residues were scored by disordered tendency higher than 0.9, indicating high probability of disordered nature of the peptide in solution. Calculated disorder tendency values are in good agreement with experimental observations and substantiate the flexibility of the free B-domain. The solution structure of the B-domain-DNA complex was reconstructed using the multi-phase ab initio modeling as is implemented in MONSA. Five individual MONSA runs resulted in similar reconstructions and χ2 values. Typical MONSA model of the B-domain in complex with DNA finished with the χ2 value 1.22 for the DNA phase (Figure 5D) and the χ2 value 1.06 for the B-domain–DNA complex phase (Figure 5E).
The MONSA model of the DNA phase was of the comparable size and shape as the molecular model of the telomeric duplex, while the protein phase formed a compact body sitting on the side of the DNA duplex. The SAXS results showed that substantial part of the unbound B-domain of TRF2 was flexible and unstructured in the solution. Upon DNA binding, the majority of the amino acids of the B-domain adopted a rigid structure and formed a compact body with Rg of ∼10 Å, diameter of ∼30 Å, and volume of ∼6000 Å3. In conclusion, SAXS measurements suggested that the B-domain of TRF2 was flexible in solution. The B-domain of TRF2 became structurally fixed upon forming the stable complex with the double stranded telomeric DNA.
DISCUSSION
The presented findings are important for understanding D-loop processing on telomeres for two main reasons.
First, the unwinding assays with BLM and RPA showed that TRF2 prevents D-loop unwinding regardless of the origin of the unwinding force. Kinetically, the binding of TRF2 to D-loop should precede DNA unwinding as both BLM and TRF2 bind DNA within seconds (30,31). The independence of the stabilizing effect on the interactions between TRF2, BLM and RPA points to a generic mechanism for the TRF2 stabilization effect.
The second important finding is that the B-domain of TRF2 became structured and rigid upon complexation with DNA. The improved rigidity of the B-domain after DNA complexation explains elegantly why TRF2 stabilizes loop structures (32), and Holliday junctions (8). The stabilization and regulation of telomeric loop structures by Rap1–TRF2 was suggested also in recent study of the Griffith laboratory describing the formation of duplex telomeric loops during the transcription (33).
Also previous studies from the Griffith and de Lange laboratories have demonstrated that TRF2 might stabilize t-loops without further specification of domain contributions. In vitro studies with the pRST5 template showed that TRF2 will generate t-loops if telomeric DNA contains a homologous 3′ G-rich single-stranded overhang (34). Furthermore, the functional elimination of TRF2 in the cell leads to a loss of t-loops, as shown by Doksani et al. using STORM imaging (32). All the beforehand mentioned studies point toward the role of full-length TRF2 and its interacting partners in forming, shaping and stabilizing of telomeric loops. More recently, Benarroch-Popivker et al. focused on the role of the central homodimerization domain of TRF2 dimer in the telomeric DNA protection (9). Their AFM experiments suggest the approximately 90 bp of DNA wrap around the TRF2 homodimerization domain. In our studies, we further described how another domain, the N-terminal B-domain of TRF2, contributes to protecting of telomeric DNA structures.
To describe solely the effect of the B-domain, without a possible interference of DNA wrapping around TRF2 homodimerization domain, we performed our studies with D-loop structures that were shorter than the length of DNA that is minimally required for DNA wrapping. Accordingly, to reveal the contribution of the B-domain to DNA binding without possible DNA wrapping effects we carried out our TRF2 binding and interaction studies with the shortest possible DNA targets (D-loops and double-stranded oligonucleotides) based on preceding studies (35,36,37). Here it is important to note that naturally occurring DNA templates in cells are significantly longer (2).
Our recent observation that TRF2 reduced D-loop unwinding disputes findings of previous studies suggesting that TRF2 stimulates DNA unwinding by BLM, with the extent dependent on TRF2 concentration (24,38). However, there are two substantial differences between our experimental setup and the approach used in previous studies: (i) we used D-loop R4 that contains the telomeric sequence; (ii) our unwinding measurements were carried out in fully saturated conditions to ensure that all binding sites on the D-loop would be occupied by TRF2. Such an arrangement might simulate molecular crowding occurring naturally at chromosome ends in cells. These conditions allowed us to observe how significant the contribution of the B-domain of TRF2 is to D-loop structure maintenance.
Moreover, the stabilization of the D-loop in the presence of full-length TRF2 and not in the presence of ΔBTRF2 was confirmed by the direct observation of thermal stability increase (Figure 3). The fluorescence based thermal stability measurement is to our knowledge the first quantitative proof that the B-domain of TRF2 is essential for the stabilization of the D-loop arrangement of telomeric DNA.
Together the stabilization of the D-loop structure and the reduction of the unwinding activity of BLM in the presence of TRF2 on telomeric DNA are in accordance with the recent study of the Yildiz laboratory (18). Bandaria et al. showed, by super-resolution microscopy, that higher recruitment of TRF2 induces a higher compacting of chromatin that might affect the access of telomerase to telomeric DNA.
Our quantifications of the impact of the B-domain on DNA binding affinity of TRF2 showed that the B-domain doubles DNA binding affinity of TRF2 compared to TRF2 protein lacking the B-domain. The increase of the binding affinity is critical in the first stage of protein-DNA interaction when a long-range electrostatic attraction takes place.
As the salt dependence of binding affinity for ΔBTRF2 and TRF2 clearly showed, the B-domain elevates the electrostatic attraction to DNA by adding at least one ion pair. The interacting ion pair most likely comprises one of the positively charged arginines or the only histidine in the B-domain (Figure 1A). In previous studies, the Gilson laboratory showed that the histidine residue in position 31 (H31) is critical for the stabilization of Holliday junctions (8). Poulet et al. have proposed that H31 contributes to the opening of double-stranded DNA. Based on our data, we were unable to specify which amino acid residue is directly responsible for the electrostatic attraction of the B-domain to DNA. However, it is also possible that the increased electrostatic attraction is a result of several partial contributions of ion pairs formed between telomeric DNA and the B-domain of TRF2, rather than a single ion pair formation.
Rap1 modulates the ability of TRF2 to bind and recognize telomeric DNA, as was documented in our recent study (16). In that study, we suggested that Rap1 increases the selectivity of TRF2 to telomeric DNA by preventing non-specific interactions of the B-domain with DNA. Thus, the removal of the B-domain from TRF2 should affect the binding to DNA similarly to complexation of TRF2 and Rap1. Indeed, we observed a similar decrease in average number of interacting DNA-protein ion pairs Z if we removed the B-domain from TRF2 (ΔZ = 1.2; Supplementary Table S1) or if we complexed Rap1 and TRF2 (ΔZ = 1.9; Supplementary Table S2 in (16)). The higher impact of Rap1 on electrostatic TRF2 attraction to DNA than the B-domain removal might be due to a steric hindrance, or allosteric and electrostatic changes induced on TRF2 upon binding of the negatively charged Rap1 (pI 4.6). Additionally, the negative charge contribution of Rap1 to the protein heterodimer might diminish long-range electrostatic attraction between the Rap1–TRF2 complex and DNA. In summary, the similar reduction of electrostatic attraction after Rap1 binding and the B-domain removal strongly supports our previous suggestion that Rap1 prevents DNA binding of the B-domain of TRF2 (16).
To shed more light on the mechanism of interaction and stabilization of DNA by the B-domain of TRF2, we described the structural arrangement of the full B-domain (44 amino acid residues) on double-stranded telomeric DNA (Figure 5 and Supplementary Table S3). SAXS measurements suggested that the B-domain was flexible in solution but became rigid upon binding to telomeric DNA. Interestingly, previous NMR measurements proposed that a peptide containing 33 amino acid residues of the B-domain remains flexible upon binding to telomeric DNA (8). The difference in flexibility could be provided by the longer sequence of the peptide used for our SAXS studies, as the formation of the firm complex of the B-domain with DNA may require the full-length B-domain. Based on our results and other available data, we speculate that the contribution of TRF2 to telomeric chromatin stabilization through non-specific interactions is mediated mainly by the B-domain. Additionally, very recent studies of the de Lange laboratory showed that TRF2 binds three-way junctions via the B-domain and thus protects t-loop cleavage (10). This finding further supports the general hypothesis that the B-domain of TRF2 is essential for stabilization of highly ordered DNA structures on telomeres. Finally, our observations that TRF2 effect on D-loop unwinding by RPA was comparable to the effect of TRF2 on D-loop unwinding by BLM helicase, suggest that the B-domain participates in a generic mechanism for stabilizing D-loop structures. The putative model for how TRF2 stabilizes D-loops via the B-domain is shown in Figure 6.
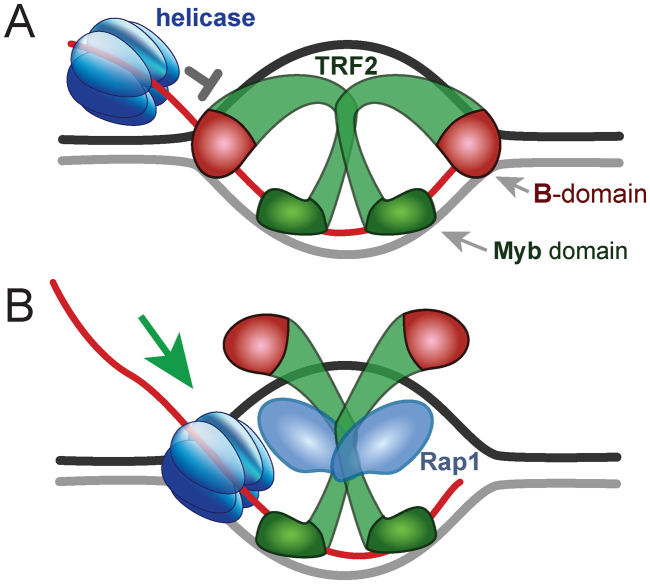
Figure 6.
Proposed mechanism for how the B-domain of TRF2 improves stability of the D-loop and how Rap1 impairs it. (A) TRF2 homodimer binds strand invasion sites of the D-loop via B-domains. Myb domains bind telomeric double-stranded regions of the D-loop. The simultaneous two-domain binding of TRF2 stabilizes the D-loop structure. (B) Rap1 prevents DNA interaction of the B-domain and thus facilitates D-loop unwinding by helicases. The relative size of DNA and protein representations is not proportional.
The B-domain of TRF2 binds branched parts of D-loops and prevents their resolution. Thus, TRF2 dimer fixes the D-loop structure via simultaneous binding of the Myb domain to double-stranded invading DNA regions and the B-domain to single-double strand junctions in the initial part of the D-loop (Figure 6A). When the complex Rap1–TRF2 binds the D-loop, Rap1 prevents the B-domain interaction with DNA. Thus, Rap1 eliminates non-specific DNA binding of TRF2 N-terminus via the B-domain. Hence, Rap1 promotes D-loop unwinding by helicases despite the TRF2 presence (Figure 6B). Please note that the size of proteins and DNA in Figure 6 does not correspond to their real relative proportions for the sake of clarity.
As the TRF2 central homodimerization domain is extremely flexible, we believe that our proposed model (Figure 6) is universal enough, to be accommodated also by more complex arrangement of telomeric duplex loops with both terminal strands inserted back into the preceding duplex (33). Furthermore, our finding that the B-domain of TRF2 improves the stability of telomeric loops is supported by a recent study of the Griffith laboratory. Kar et al. suggested that Rap1–TRF2 may play a role in controlling the stability of D-loop structures (33).
Latest studies show the ability of the B-domain of TRF2 to interact directly with histones in vitro and in vivo (39). Such bi-functional (DNA-and-protein) binding of the B-domain would enable TRF2 to interconnect tightly telomeric DNA and histones. Thus, the net of condensing interactions of TRF2 within chromatin might contribute to more interconnected and better-protected human chromosome ends. The chromosome tightening ability of TRF2 might be modified by Rap1 binding. Rap1 and TRF2 complexing diminish DNA binding through the B-domain. Accordingly, Rap1 may neglect nonspecific DNA interactions of TRF2 via the B-domain and thus mediate loosening of chromatin when it is necessary during the telomere extension and chromosome end replication.
In summary, we showed that the B-domain is critical for the ability of TRF2 to reduce D-loop unwinding regardless of the cause of unwinding. We revealed that Rap1 complexation with TRF2 likely prevents interaction of the B-domain of TRF2 with the D-loop structure and thus counteracts B-domain effects on the DNA binding properties of TRF2. Additionally, we quantified the stabilization of the telomeric D-loop upon TRF2 binding. Our DNA binding analysis of TRF2 and ΔBTRF2 showed that the B-domain contributes to overall DNA binding affinity significantly, mainly via improved nonspecific electrostatic interactions. We further revealed that the full B-domain of TRF2 became rigid upon binding to double stranded DNA. The improved rigidity of the bound B-domain contributed to overall D-loop stability. We showed that Rap1, upon binding TRF2, lessens the B-domain interaction with DNA. Based on our quantitative analyses, we proposed the mechanism for how TRF2 and Rap1 regulate telomeric D-loop unwinding.
DATA AVAILABILITY
All SAXS datasets and MONSA models have been deposited to the Small Angle Scattering Biological Data Bank as entries: SASDC62, SASDB79 and SASDB89.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Titia de Lange and Thomas Cech for their encouraging discussion and suggestions provided during CSHL conference Telomeres and Telomerase 2017, Michal Zimmerman for ΔBTRF2 cloning constructs preparations, Jiří Fajkus for generous scientific and personal support in all circumstances, Mate Gyimesi for providing BLM construct, Victoria Marini for her advice during helicase assays, Mario Špírek for his assistance in RPA preparation and Richard Štefl for providing us with pET28bSMT3 vector. We are grateful to Simon Boulton for his igniting indication of the B-domain involvement in D-loop stabilization, practical advice and long lasting collegial encouragement. We are indebted to the colleagues from Biomolecular Interaction and Crystallization and Proteomic core facilities of CEITEC for their excellent assistance. We are grateful to Victoria Marini for the critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation [16–20255S to C.H.]; Ministry of Education, Youth and Sports of the Czech Republic (CEITEC 2020 project) [LQ1601]; European Union’s Horizon 2020 Research and Innovation Program [692298]. Funding for open access charge: Czech Science Foundation [16–20255S].
Conflict of interest statement. None declared.
REFERENCES
- 1. Greider C.W., Blackburn E.H.. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985; 43:405–413. [DOI] [PubMed] [Google Scholar]
- 2. Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T.. Mammalian telomeres end in a large duplex loop. Cell. 1999; 97:503–514. [DOI] [PubMed] [Google Scholar]
- 3. Greider C.W. Telomeres do D-loop–T-loop. Cell. 1999; 97:419–422. [DOI] [PubMed] [Google Scholar]
- 4. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 5. Nandakumar J., Cech T.R.. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013; 14:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feuerhahn S., Chen L.Y., Luke B., Porro A.. No DDRama at chromosome ends: TRF2 takes centre stage. Trends Biochem. Sci. 2015; 40:275–285. [DOI] [PubMed] [Google Scholar]
- 7. Court R., Chapman L., Fairall L., Rhodes D.. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005; 6:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poulet A., Buisson R., Faivre-Moskalenko C., Koelblen M., Amiard S., Montel F., Cuesta-Lopez S., Bornet O., Guerlesquin F., Godet T. et al. . TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009; 28:641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benarroch-Popivker D., Pisano S., Mendez-Bermudez A., Lototska L., Kaur P., Bauwens S., Djerbi N., Latrick C.M., Fraisier V., Pei B. et al. . TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Mol. Cell. 2016; 61:274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmutz I., Timashev L., Xie W., Patel D.J., de Lange T.. TRF2 binds branched DNA to safeguard telomere integrity. Nat. Struct. Mol. Biol. 2017; 24:734–742. [DOI] [PubMed] [Google Scholar]
- 11. Saint-Leger A., Koelblen M., Civitelli L., Bah A., Djerbi N., Giraud-Panis M.J., Londono-Vallejo A., Ascenzioni F., Gilson E.. The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres. Cell Cycle. 2014; 13:2469–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Croteau D.L., Popuri V., Opresko P.L., Bohr V.A.. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014; 83:519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarek G., Vannier J.B., Panier S., Petrini J.H.J., Boulton S.J.. TRF2 recruits RTEL1 to telomeres in S phase to promote T-loop unwinding. Mol. Cell. 2015; 57:622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sarkar J., Wan B.B., Yin J.H., Vallabhaneni H., Horvath K., Kulikowicz T., Bohr V.A., Zhang Y.B., Lei M., Liu Y.. SLX4 contributes to telomere preservation and regulated processing of telomeric joint molecule intermediates. Nucleic Acids Res. 2015; 43:5912–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Georgaki A., Strack B., Podust V., Hubscher U.. DNA unwinding activity of replication protein A. FEBS Lett. 1992; 308:240–244. [DOI] [PubMed] [Google Scholar]
- 16. Janouskova E., Necasova I., Pavlouskova J., Zimmermann M., Hluchy M., Marini V., Novakova M., Hofr C.. Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res. 2015; 43:2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sarthy J., Bae N.S., Scrafford J., Baumann P.. Human RAP1 inhibits non‐homologous end joining at telomeres. EMBO J. 2009; 28:3390–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bandaria JN, Jigar N., Qin P., Berk V., Chu S., Yildiz A.. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell. 2016; 164:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Busso D., Delagoutte-Busso B., Moras D.. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 2005; 343:313–321. [DOI] [PubMed] [Google Scholar]
- 20. Youds J.L., Mets D.G., McIlwraith M.J., Martin J.S., Ward J.D., ONeil N.J., Rose A.M., West S.C., Meyer B.J., Boulton S.J.. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010; 327:1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smogorzewska A., Van Steensel B., Bianchi A., Oelmann S., Schaefer M.R., Schnapp G., De Lange T.. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000; 20:1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 1996; 237:260–273. [DOI] [PubMed] [Google Scholar]
- 23. Hofr C., Sultesova P., Zimmermann M., Mozgova I., Schrumpfova P.P., Wimmerova M., Fajkus J.. Single-Myb-histone proteins from Arabidopsis thaliana: a quantitative study of telomere-binding specificity and kinetics. Biochem. J. 2009; 419:221–228. [DOI] [PubMed] [Google Scholar]
- 24. Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A.. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002; 277:41110–41119. [DOI] [PubMed] [Google Scholar]
- 25. Sigurdsson S., Trujillo K., Song B.W., Stratton S., Sung P.. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 2001; 276:8798–8806. [DOI] [PubMed] [Google Scholar]
- 26. Janscak P., Garcia P.L., Hamburger F., Makuta Y., Shiraishi K., Imai Y., Ikeda H., Bickle T.A.. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J. Mol. Biol. 2003; 330:29–42. [DOI] [PubMed] [Google Scholar]
- 27. Opresko P.L., Otterlei M., Graakjær J., Bruheim P., Dawut L., Kølvraa S., May A., Seidman M.M., Bohr V.A.. The werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004; 14:763–774. [DOI] [PubMed] [Google Scholar]
- 28. Georgaki A., Strack B., Podust V., Hubscher U.. DNA unwinding activity of replication protein-A. FEBS Lett. 1992; 308:240–244. [DOI] [PubMed] [Google Scholar]
- 29. Dosztanyi Z., Csizmok V., Tompa P., Simon I.. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005; 347:827–839. [DOI] [PubMed] [Google Scholar]
- 30. Mattern K.A., Swiggers S.J.J., Nigg A.L., Lowenberg B., Houtsmuller A.B., Zijlmans J.M.J.M.. Dynamics of protein binding to telomeres in living cells: Implications for telomere structure and function. Mol. Cell. Biol. 2004; 24:5587–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang S., Qin W., Li J.-H., Lu Y., Lu K.-Y., Nong D.-G., Dou S.-X., Xu C.-H., Xi X.-G., Li M.. Unwinding forward and sliding back: an intermittent unwinding mode of the BLM helicase. Nucleic Acids Res. 2015; 43:3736–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doksani Y., Wu J.Y., de Lange T., Zhuang X.. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013; 155:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kar A., Willcox S., Griffith J.D.. Transcription of telomeric DNA leads to high levels of homologous recombination and t-loops. Nucleic Acids Res. 2016; 44:9369–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stansel R.M., de Lange T., Griffith J.D.. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001; 20:5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bilaud T., Koering C.E., BinetBrasselet E., Ancelin K., Pollice A., Gasser S.M., Gilson E.. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996; 24:1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Broccoli D., Smogorzewska A., Chong L., de Lange T.. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997; 17:231–235. [DOI] [PubMed] [Google Scholar]
- 37. Hanaoka S., Nagadoi A., Yoshimura S., Aimoto S., Li B., de Lange T., Nishimura Y.. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J. Mol. Biol. 2001; 312:167–175. [DOI] [PubMed] [Google Scholar]
- 38. Lillard-Wetherell K., Machwe A., Langland G.T., Combs K.A., Behbehani G.K., Schonberg S.A., German J., Turchi J.J., Orren D.K., Groden J.. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004; 13:1919–1932. [DOI] [PubMed] [Google Scholar]
- 39. Konishi A., Izumi T., Shimizu S.. TRF2 protein interacts with core histones to stabilize chromosome ends. J. Biol. Chem. 2016; 291:20798–20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All SAXS datasets and MONSA models have been deposited to the Small Angle Scattering Biological Data Bank as entries: SASDC62, SASDB79 and SASDB89.