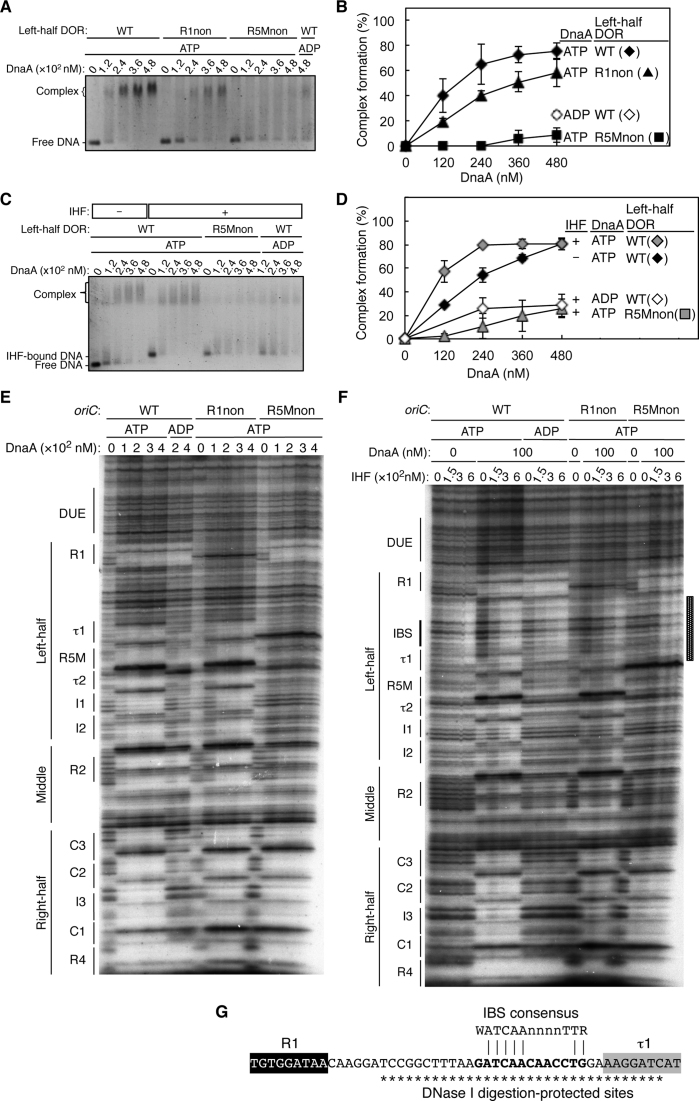
Figure 2.
DnaA bound to R5M, but not R1, is essential for DnaA assembly on the left-half DOR. (A–D) EMSA with the left-half DOR bearing R1non or R5Mnon mutations. Each DNA fragment (35 nM) was incubated with the indicated amount of DnaA in the absence (A) or presence (C) of 72 nM IHF, then analyzed by 2% agarose-gel electrophoresis. The amount of DNA complexed with DnaA relative to the input DNA is shown as ‘Complex formation (%)’ in (B) and (D). Two independent experiments were carried out, and both data, mean values, and representative gel images in a black-white inverted mode are shown. (E–G) DNase I-footprint assay with oriC fragments (346 bp) containing R1non or R5Mnon mutations. Indicated amounts of ATP–DnaA or ADP–DnaA were incubated for 10 min at 30°C with 2.4 nM 32P-end-labeled oriC and 3 mM ATP or ADP in the absence (E) or presence (F) of 150–600 nM IHF, followed by DNase I digestion and gel-electrophoresis analysis. Positions of DNA motifs are indicated. The sequence of the region protected by IHF binding is shown in (G). The IBS consensus sequence and DNase I digestion-protected sites are indicated by bold lettering and asterisks, respectively.