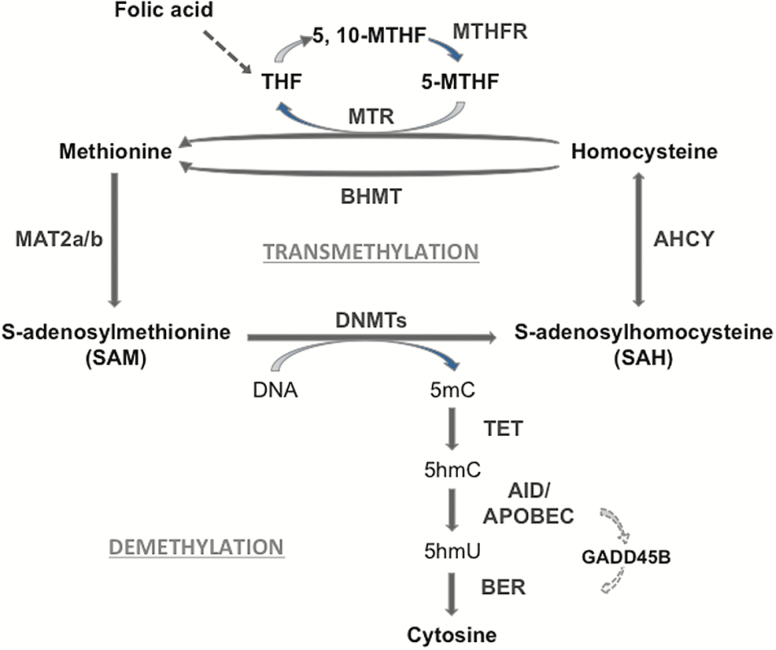
Figure 1.
One-carbon metabolism, transmethylation reactions and DNA demethylation. DNA methylation on specific cytosine moieties is catalyzed by DNA-methyltransferases (DNMTs). This reaction is dependent on the levels of S-adenosyl-methionine (SAM), which is synthesized in the brain by the methionine-adenosyl-transferase (MAT) 2. After the DNA methyl transfer reaction, S-adenosyl-homocysteine (SAH) is formed as a by-product. SAH exerts a feedback inhibitory activity on DNMT and is used as a substrate for the adenosyl homocysteine hydrolase (AHCY) leading to the synthesis of homocysteine. The remethylation of homocysteine to form methionine is catalyzed via the folate cycle, that is, the conversion of folic acid into tetrahydrofolate (THF) and 5,10-methylenete-trahydrofolate (5,10-MTHF). The methylene tetrahydrofolate reductase (MTHFR) will produce 5-methyltetrahydrofolate (5-MTHF) that is used as a substrate by methionine synthase (MTR) to form methionine. The synthesis of methionine may also involve the activation of the betaine homocysteine methyltransferase (BHMT) pathway. DNA demethylation is initiated by ten-eleven-translocase (TET) enzymes that hydroxylate 5-methylcytosine (5mC) forming 5-hydroxymethylcytosine (5hmC). In the end, through a GADD45B coordinated process, 5hmC is deaminated to 5-hydroxyuracil (5hmU) by cytidine deaminases (i.e., AID/APOBECs). The base excision repair (BER) pathway removes 5hmU and substitutes it with cytosine.