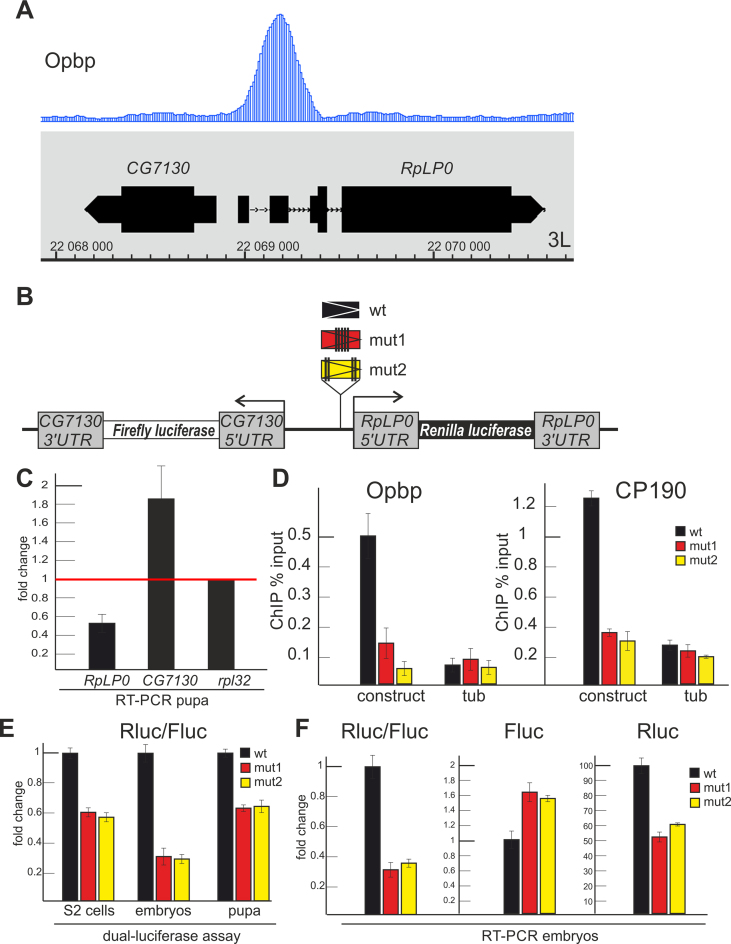
Figure 4.
Opbp regulates the RplP0 promoter in transgenic flies. (A) Track for Opbp binding profile is presented at the CG7130/RplP0 genome region. (B) Scheme of transgenic constructs used to examine the role of Opbp (wt and two mutants of Opbp motifs) at the RplP0 promoter. (C) Changes of expression levels of RplP0 and CG7130 measured by RT-qPCR with cDNAs synthesized from RNA extracted from wt and opbp− pupa. Individual transcript levels were determined by RT-qPCR with corresponding primers normalized relative to RpL32 for the amount of input cDNA. Histogram shows the changes of RplP0 and CG7130 relatively expression levels in wild type pupa (red line at the ‘1’ on the scale). Error bars show standard deviation of triplicate PCR measurements. (D) Both Opbp and CP190 are recruited to the transgenic construct containing the Opbp motif. ChIP-real-time-PCR of Opbp and CP190 to the transgenic constructs; percentage of input recovery is shown. Error bars indicate standard deviations of quadruplicate PCR measurements for three independent experiments. (E), (F) Opbp/CP190 recruitment is required for reporter expression driven by the RplP0 promoter. Histogram shows the ratio of Renilla to Firefly luciferases in S2 cells (left) and transgenic fly lines (right) in dual-luciferase assay (E) and relative amount of luciferase mRNAs (F) in transgenic fly lines.