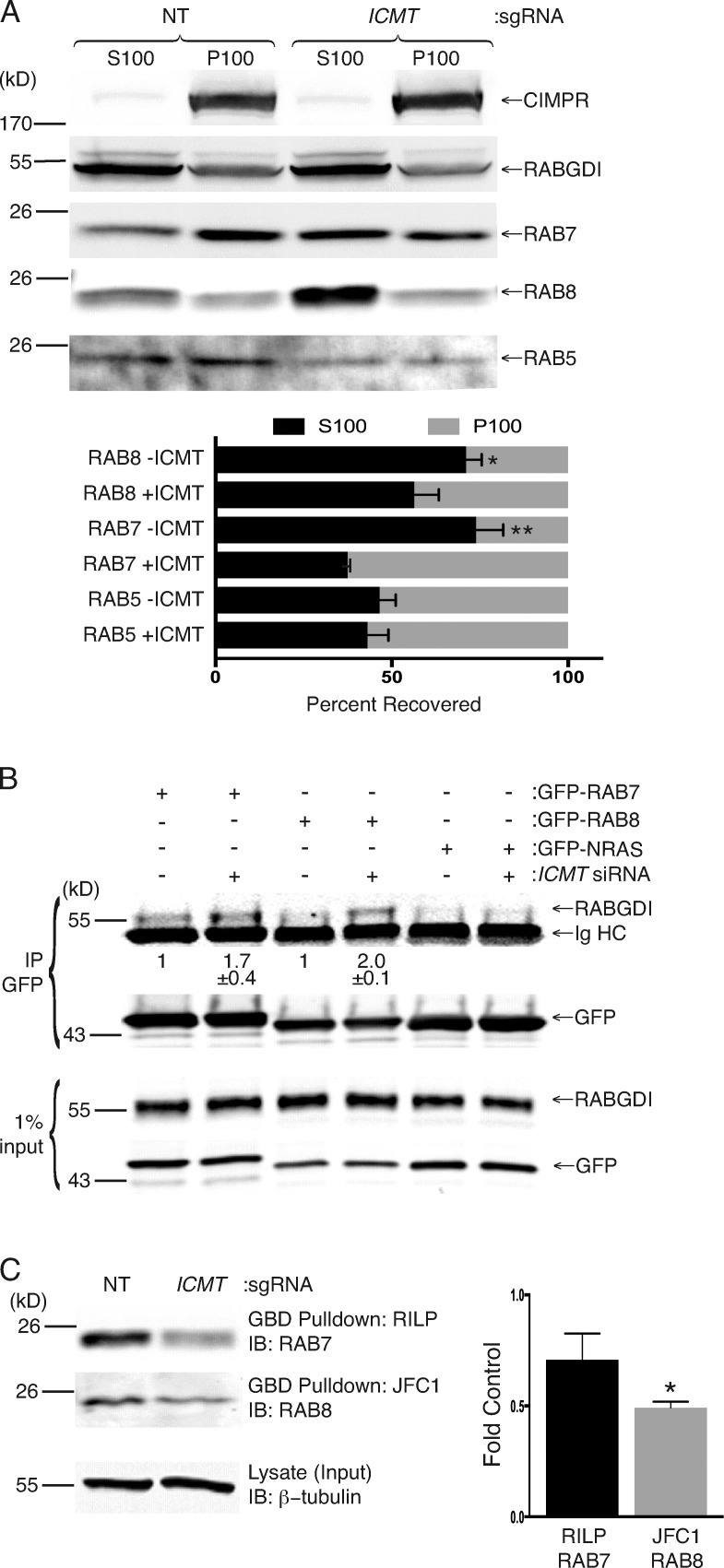
Figure 4.
Unmethylated RAB7 and RAB8 are enriched in the cytosol, where they interact with the cytosolic chaperone RABGDI, and GTP loading is decreased. (A) Cell equivalents of cytosolic (S100) and membrane (P100) fractions generated from SKMEL-28 cells disrupted by nitrogen cavitation with or without genomic disruption of ICMT by CRISPR/Cas9 were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. The graph under the representative immunoblots (IBs) shows means ± SEM of the percentage of endogenous RAB5, RAB7, and RAB8 present in each fraction with or without ICMT CRISPR (n = 3). (B) HEK293 cells were treated for 3 d with or without NT or ICMT siRNA before transfection with the indicated GFP constructs. The following day, the cells were lysed, the GFP-tagged proteins were immunoprecipitated (IP), and the precipitates and input were immunoblotted for RABGDI and GFP. Values shown under the representative blot are means ± SEM of the amount of RABGDI coimmunoprecipitated with GFP-RAB7 and GFP-RAB8 normalized to the input as well as the amount immunoprecipitated without ICMT siRNA (n = 3). (C) GTP loading of RAB7 and RAB8 in SKMEL-28 cells with or without genomic disruption of ICMT. GTP-bound RAB7 was quantified by GST-RILP pulldown, and GTP-RAB8 was quantified by GST-JFC1 pulldown. Graphs show the amount of GTP-RAB7 and GTP-RAB8 normalized to the loading control. n = 3. *, P < 0.05; **, P < 0.01 (two-sided t test).