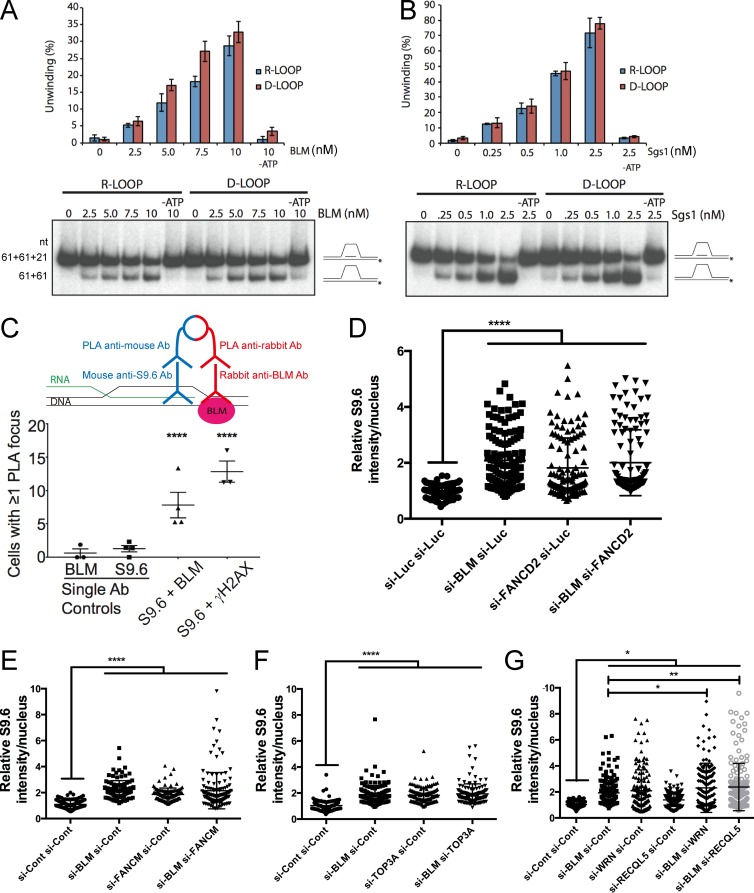
Figure 7.
Potential mechanism of BLM-dependent R-loop mitigation. Comparative R-loop and D-loop unwinding by BLM (A) and Sgs1 (representive gel shown of three separate experiments is shown; error bars are SEM; B). Schematics of the slow migrating oligonucleotide loop substrate and faster migrating product are shown at right and nucleotide (nt) sizes are listed to the left of A. Protein concentrations used are listed above. Quantification of unwinding efficiency is shown in the graph. (C) Proximity ligation of BLM and DNA:RNA hybrids in cells. A schematic of PLA (above) shows that signal only emerges when two epitopes (DNA:RNA and BLM) are close enough to ligate oligonucleotides conjugated to secondary antibodies (Ab). S9.6 and γ-H2AX were previously associated in cells using PLA and serve as a positive control (Stork et al., 2016). Cells with any fluorescence signal were scored as positive. Pooled count data for single primary antibody controls and dual antibody PLA reactions were compared using a Fisher’s exact test. (D–G) Nuclear S9.6 staining data for the indicated siRNA treatments. Dot plots show the range of values quantified. Western blots to confirm double knockdown efficiency are shown in Fig. S3. P-values were determined by ANOVA with Sidak’s multiple comparisons test post hoc. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.