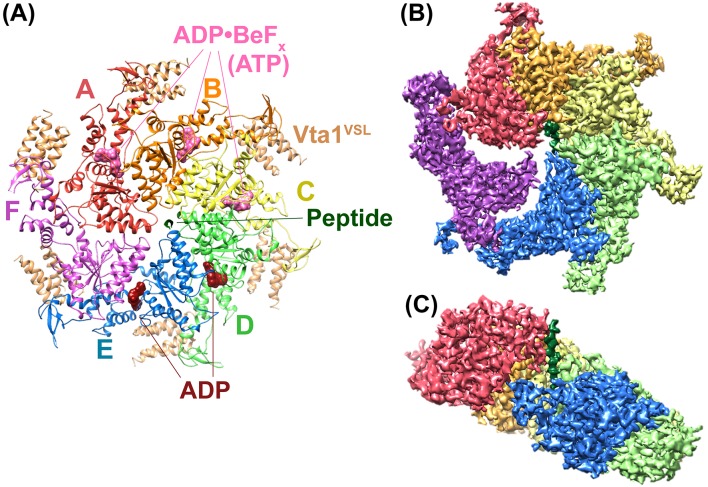
Figure 1. Overall structure of the Vps4 complex.
(A) Ribbon representation of the complex viewed from the ‘top’ N-terminal side of Vps4 and N-terminal end of the peptide. (B) Similar orientation as panel A showing a segmented map contoured around Vps4 and peptide. (C) Same as panel B viewed from the side with density for subunit F removed for clarity.
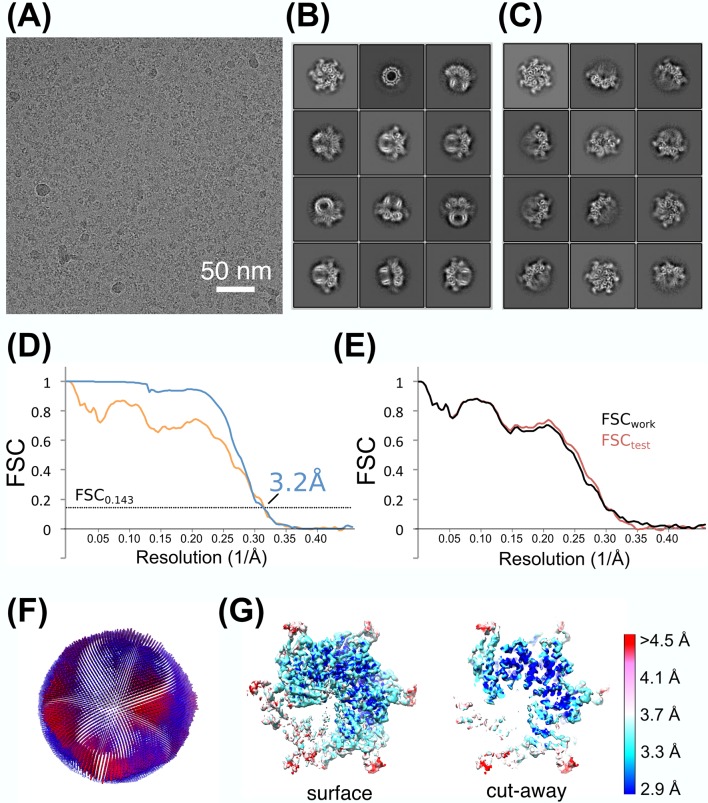
Figure 1—figure supplement 1. Cryo-EM of the Vps4 complex.
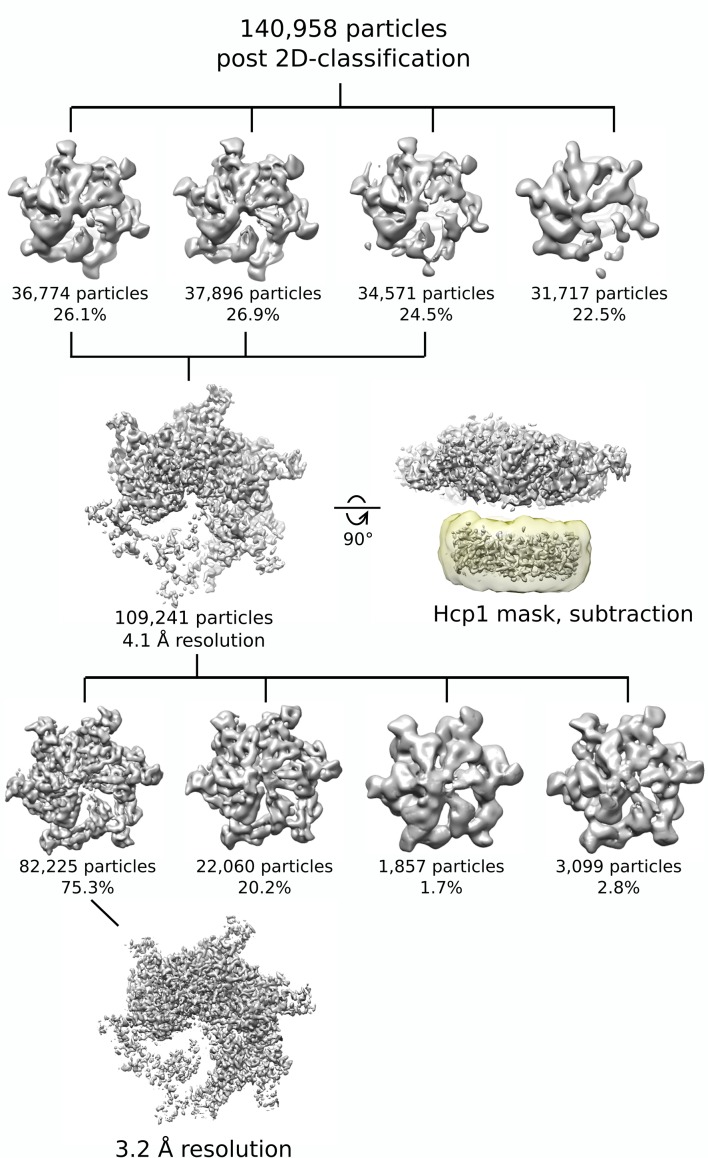
Figure 1—figure supplement 2. Classification and signal-subtraction scheme for the Vps4 complex.
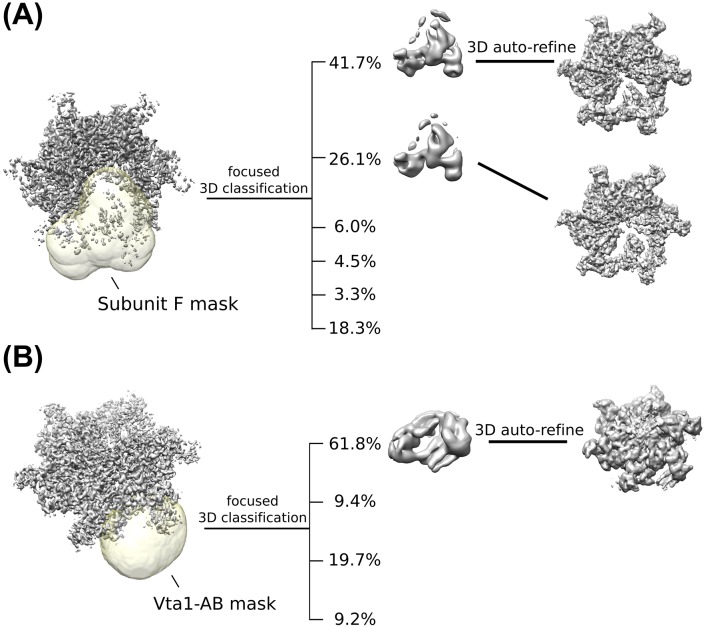
Figure 1—figure supplement 3. Focused classification of Vps4 subunit F and Vta1.
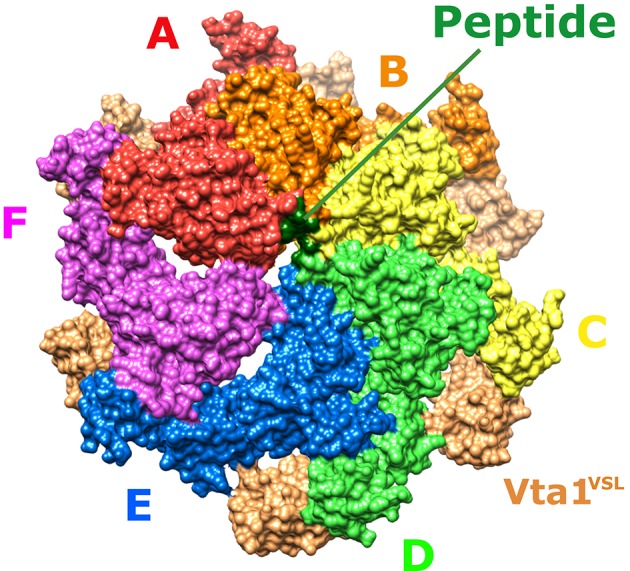
Figure 1—figure supplement 4. Surface Representation.