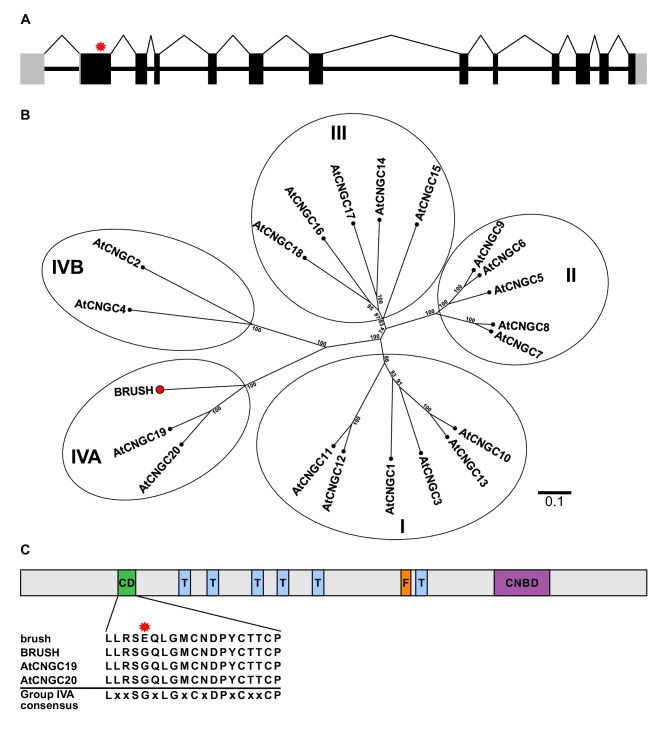
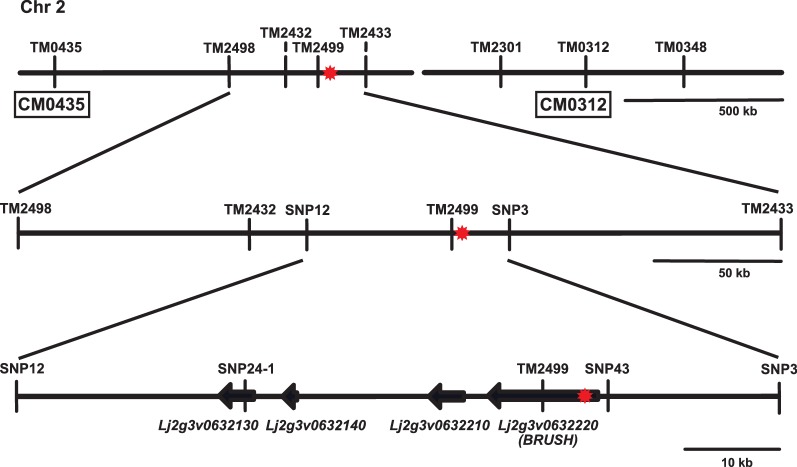
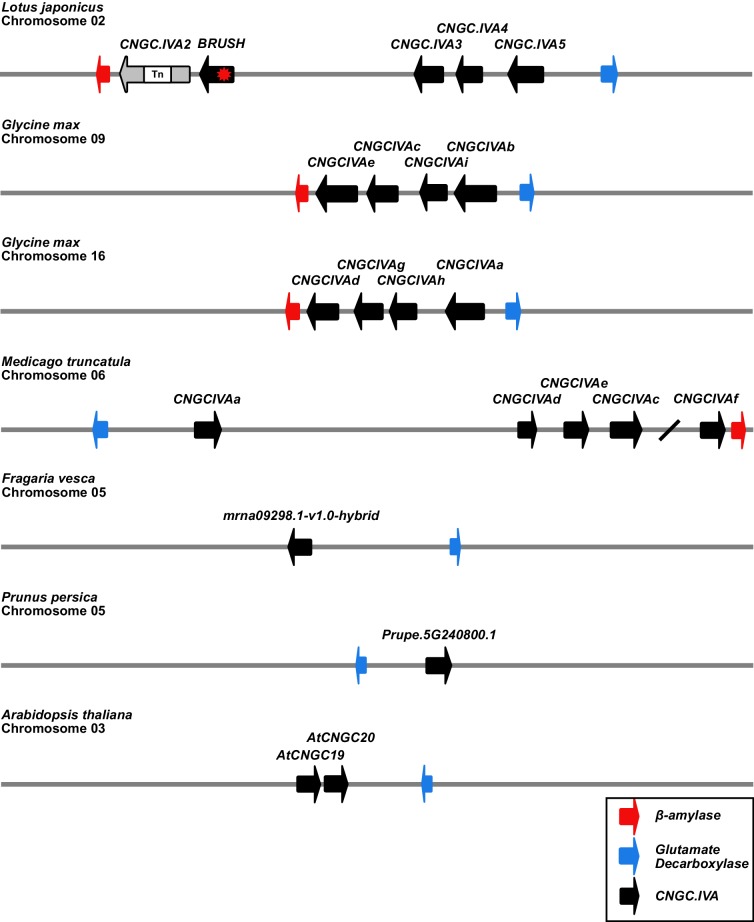
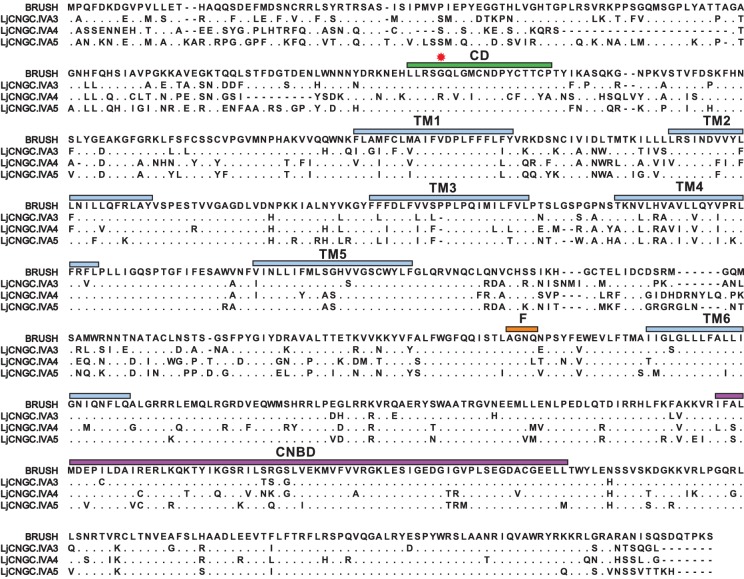
Figure 1. brush contains a point mutation in CNGC.IVA1 (BRUSH).
(A) Schematic of the intron-exon structure of BRUSH highlighting the brush mutation in the first exon (red asterisk). (B) Phylogenetic tree of BRUSH (red node end) in relation to Arabidopsis thaliana CNGC proteins. BRUSH is orthologous to the Group IVA members AtCNGC19 and AtCNGC20. (C) Overview of the BRUSH protein domain structure highlighting the conserved Group IVA domain (CD, green), transmembrane domains (T, light blue), putative filter region (F, orange), and the predicted cyclic nucleotide-binding domain (CNBD, purple). Shown below is the CD sequence in brush (G134E mutation, asterisk) relative to BRUSH, AtCNGC19, and AtCNGC20 and the Group IVA CNGC consensus (Yuen and Christopher, 2013). Numbers at the branch points in (B) indicate the percentage bootstrap values (100 iterations) for the inferred tree. Scale bar in (B) indicates the number of amino acid substitutions per site.