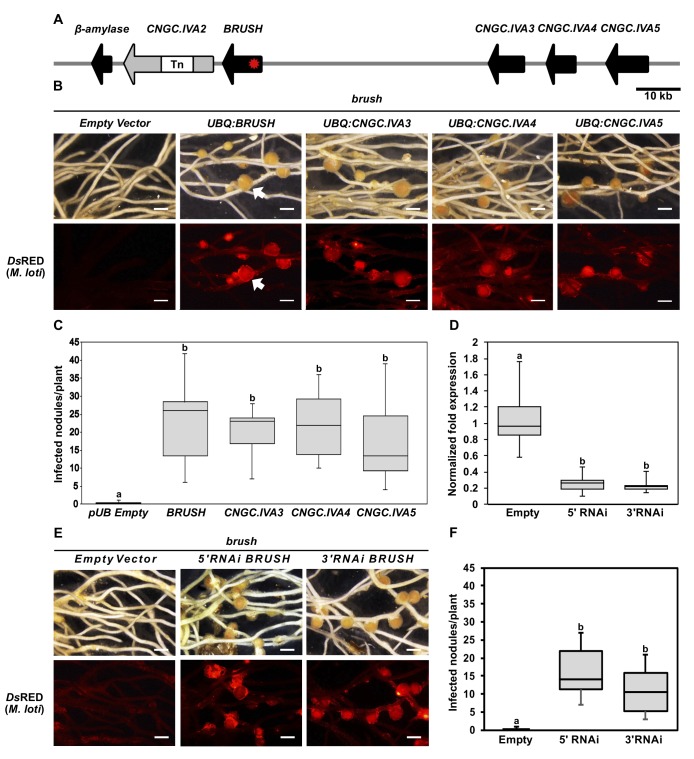
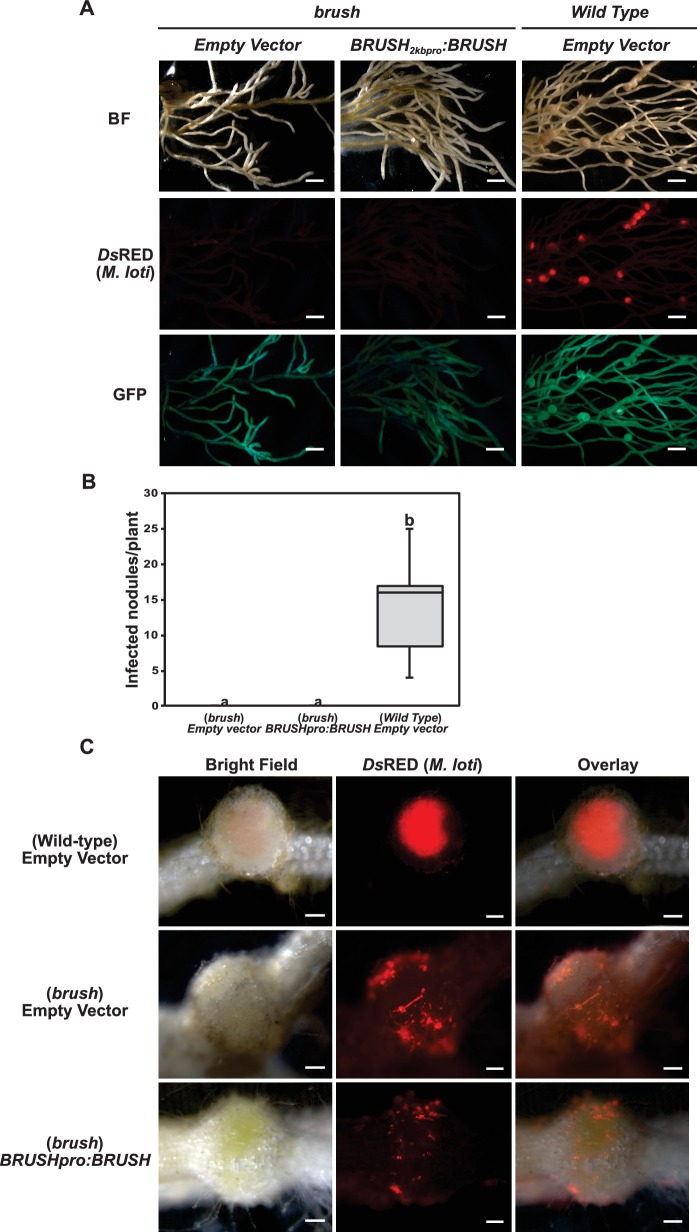
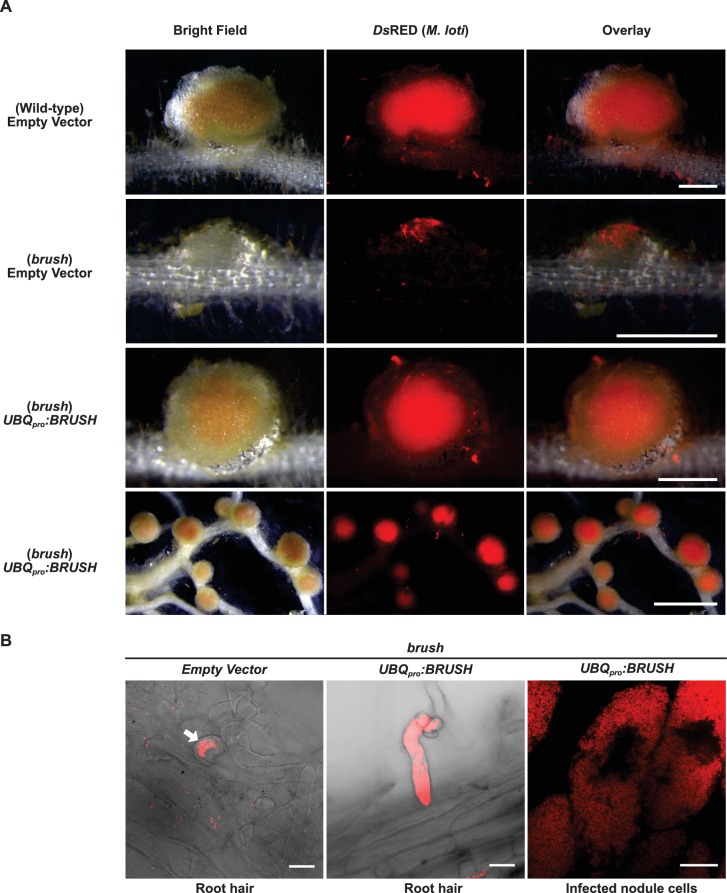
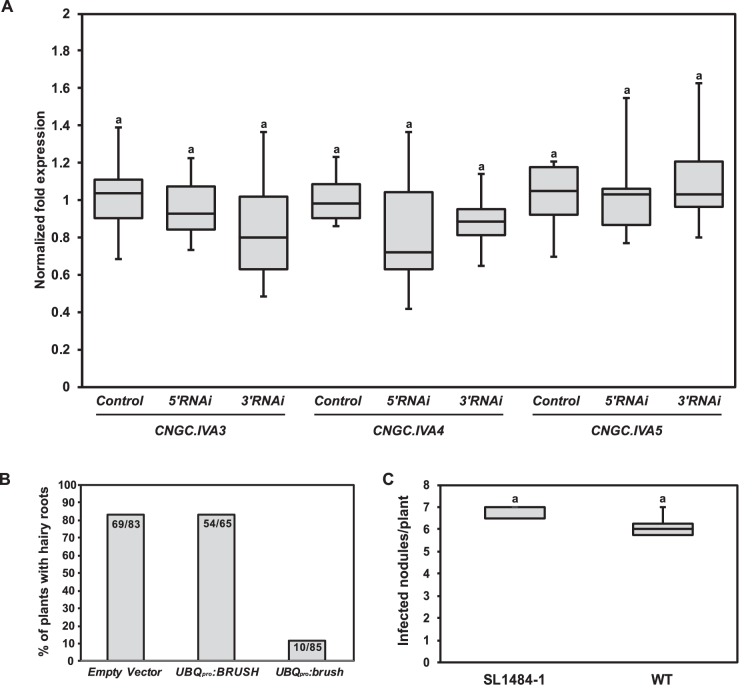
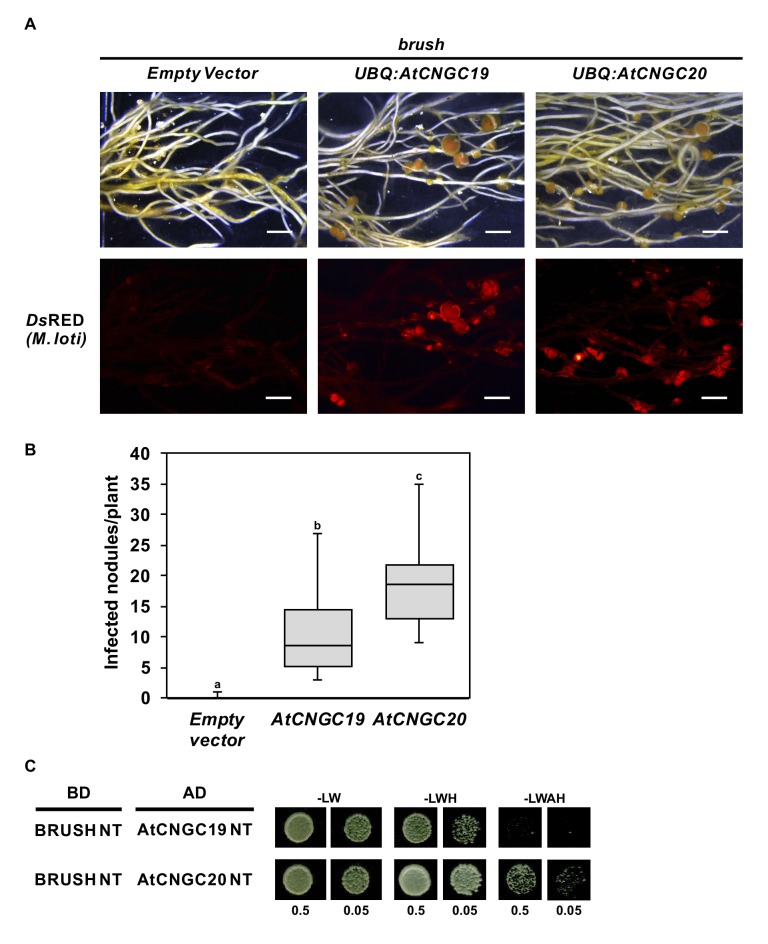
Figure 2. Genetic complementation of brush.
(A) Genomic region surrounding BRUSH on L. japonicus chromosome 2 showing a cluster of five CNGC.IVA genes (red asterisk, brush causative mutation). CNGC.IVA2 contains a large transposon (Tn) insertion. CNGC.IVA2 contains a large transposon insertion (shown as a gap). (B) Complementation assay of brush roots by overexpression (ubiquitin promoter, UBQ) of the four expressed CNGC.IVA cluster members (bright field, top panel). The presence of red fluorescent nodules (arrow) colonized by Mesorhizobium loti expressing the red fluorescent protein DsRED (lower panel) indicates successful bacterial infection and thus complementation. (C) Number of nodules per transformed plant from (B) (n = 10 for all constructs). (D) Quantitative reverse-transcription PCR analysis of brush transcript levels after RNAi targeting either the 5'UTR or 3' UTR of brush. The normalized fold expression of brush is shown relative to empty vector control roots (n = 6 for all constructs). (E) Complementation analysis of brush expressing RNAi fragments targeting either the 5'UTR or 3'UTR of brush in the brush mutant. Panels are the same as (B). (F) Number of nodules per transformed plant (n = 10 for all constructs) from (E). Roots for both complementation experiments were observed 6 weeks after inoculation with rhizobia. Scale bars in (B) and (E) represent 1 mm. Letters in (C), (D), and (F) indicate different statistical groups (ANOVA followed by Tukey’s HSD test). F(4, 45)=10.86, p < 0.001 (C), F(2, 15)=20.82, p < 0.001 (D), F(2, 27)=22.72, p < 0.001 (F).