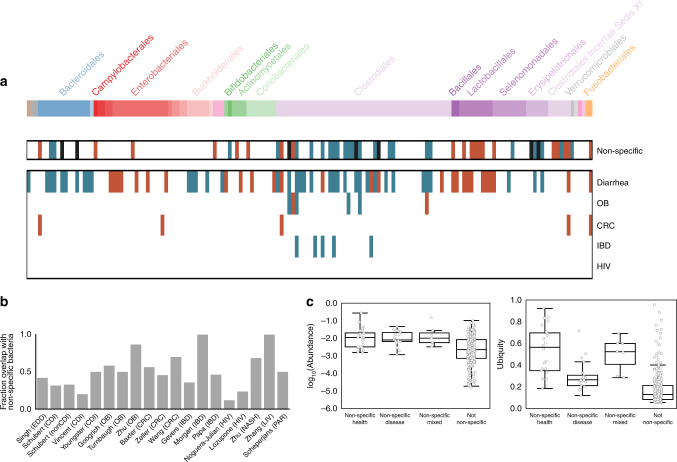
Fig. 3.
The majority of disease-associated microbiome associations overlap with a non-specific microbial response to disease. a Non-specific and disease-associated genera. Genera are in columns, arranged phylogenetically according to a PhyloT tree built from genus-level NCBI IDs (http://phylot.biobyte.de). Non-specific genera are associated with health (or disease) in at least two different diseases (q < 0.05, KW test, Benjamini–Hochberg FDR correction). Disease-specific genera are significant in the same direction in at least two studies of the same disease (q < 0.05, FDR KW test). As in Fig. 2, blue indicates higher mean abundance in controls and red indicates higher mean abundance in patients. Black bars indicate mixed genera which were associated with health in two diseases and also associated with disease in two diseases. Disease-specific genera are shown for diseases with at least three studies. Phyla, left to right: Euryarchaeota (brown), Verrucomicrobia Subdivision 5 (gray), Candidatus Saccharibacteria (gray), Bacteroidetes (blue), Proteobacteria (red), Synergistetes (pink), Actinobacteria (green), Firmicutes (purple), Verrucomicrobia (gray), Lentisphaerae (pink), Fusobacteria (orange). See Supplementary Fig. 3 for genus labels. b The percent of each study’s genus-level associations which overlap with the shared response (q < 0.05, FDR KW test). Only data sets with at least one significant association are shown. c Overall, abundance and ubiquity of non-specific genera across all patients in all data sets. Non-specific genera on the x-axis are as defined above