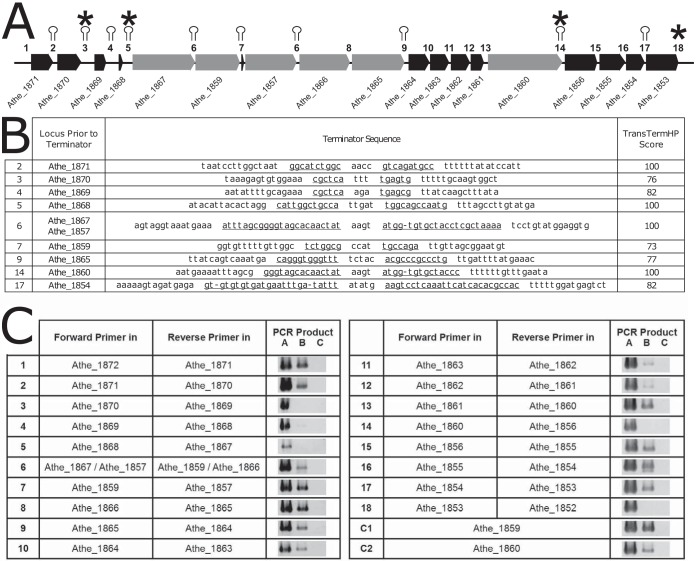
FIG 2.
In silico and experimental examination of the GDL operon structure. (A) Structure of the C. bescii GDL, with in silico terminator predictions shown as stem-loops and PCR-determined terminators marked with asterisks. Intergenic regions are numbered 1 to 18. Note that intergenic region 6 appears twice because the DNA sequence for Athe_1867 to Athe_1859 is identical to that for Athe_1857 to Athe_1866 (see the discussion of Fig. 3 in the text). (B) In silico terminator sequence predictions corresponding to intergenic regions 1 to 18 in the GDL of C. bescii, as predicted by TransTermHP (32). The complementary sequence of each stem-loop is underlined, and the TransTermHP score is shown. (C) Experimental verification of terminator sequences by PCRs targeting the regions between genes. Forward primers were designed to bind in one gene, with the reverse primer binding in the next sequential gene (see Table S2 in the supplemental material) to amplify through intergenic regions 1 to 18. PCR A was a positive control using C. bescii genomic DNA as the template. PCR B used reverse-transcribed RNA as the template. PCR C was a negative control on non-reverse-transcribed RNA to verify that there was no DNA contamination in the isolated RNA sample. A band for PCR B indicates that there is an mRNA stretching between the two genes to which the forward and reverse primers bind. Two positive-control reactions (C1 and C2) were designed, with both the forward and reverse primers targeting a single gene (Athe_1859 or Athe_1860).