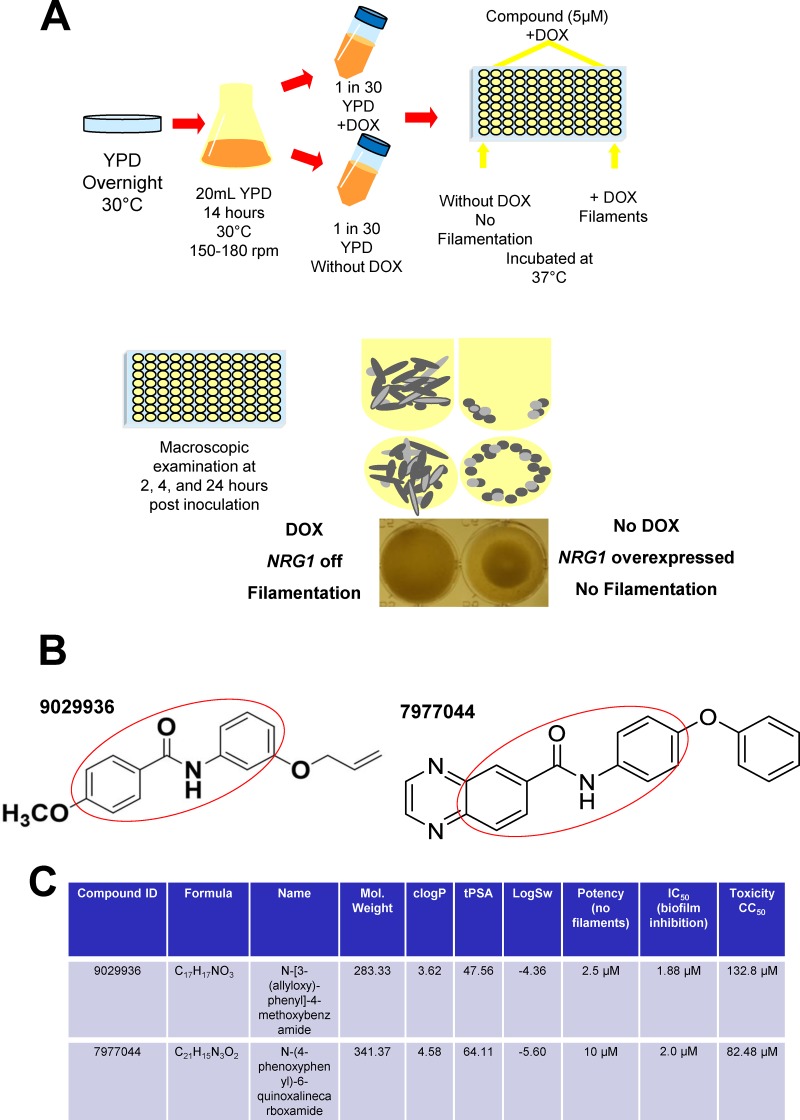
FIG 1 .
Primary screen to identify inhibitors of C. albicans filamentation. (A) Schematic diagram of the phenotypic assay used for large-scale phenotypic screening of 30,000 small-molecule compounds in the DIVERSet chemical library, which we used in our search for inhibitors of C. albicans filamentation. The screening uses 96-well round-bottom microtiter plates and takes advantage of tight control via doxycycline of morphogenetic conversions in the C. albicans tet-NRG1 strain. Individual wells of the microtiter plates are seeded with fungal cells in the presence of 5 µM each individual compound, with appropriate positive and negative controls. The plates are incubated at 37°C and visually inspected at 2 h, 4 h, and 24 h. Under the conditions used, wells containing cells that grow the filamentous form (uninhibited by the presence of the compound) appear cloudy, whereas cells that grow in the yeast form (due to inhibition of filamentation in the presence of a hit compound) fall to the bottom of the wells and form rings, which are easily discernible macroscopically. Microscopy is then used for confirmation of the inhibitory effect on filamentation. (B) Chemical structures of compounds 9029936 and 7977044, two of the major initial hits identified in the primary screen. The two compounds share a common biaryl amide motif (highlighted in red). (C) Identity, physicochemical properties, including the clogP (partition coefficient and a measure of lipophilicity), tPSA (molecular polar surface area), and logSw (solubility of the drug in water), as well as IC50 (potency) and CC50 (toxicity) values, for these two small-molecule compounds.