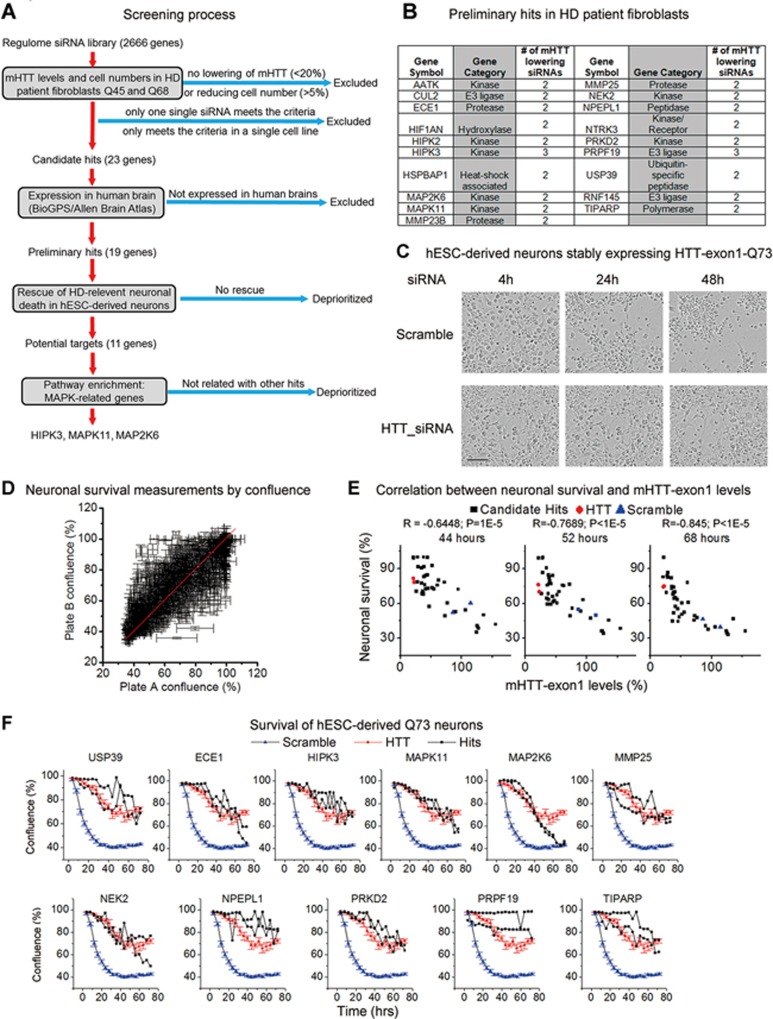
Figure 1.
Potential druggable genetic modifiers of mHTT levels identified by screening. (A) A schematic flowchart showing the screening process. (B) The information of potential preliminary hits. mHTT levels were measured by HTRF using the 2B7/MW1 antibody pair in two different human HD patient fibroblast lines (Q68 versus Q45). “# of mHTT lowering siRNAs” indicates the number of siRNAs (out of four) that reduce mHTT levels in both lines. (C) When cultured under standard (non-protective) conditions, hESC-derived neurons stably expressing HTT-exon1 fragments exhibited a long polyQ (Q73) specific degeneration phenotype, which could be assessed by imaging-based measurements. Representative images (of over 20 biological replicates) show the neuronal survival changes in these cells (Q73 neurons) transfected with scrambled siRNA (scramble) or HTT-exon1 siRNA (HTT). Scale bar, 50 μm. (D) The candidate hits were tested in Q73 neurons using SMART-pool siRNAs (Dharmacon) and the confluences measured by imaging at different time points were calculated by IncuCyte based on four fields in each well. The signals from two independent transfections (Plate A versus B) show consistency, signals are clustered near the diagonal line (red). (E) In Q73 neurons transfected with siRNA-pools targeting the primary hits, the averaged confluences of wells transfected with each siRNA in each plate were plotted with the mHTT-exon1 levels (eight measurements from two independent transfected wells) and measured by HTRF using the 2B7/MW1 antibody pair 48 h after transfection. The correlation coefficient R and P values for confluences measured at 68 h were calculated by Spearman correlation analysis. (F) Neuronal survival plots of Q73 neurons transfected with indicated siRNAs. Neuronal survival was measured by the averaged confluence of four fields in each well. Note that the scramble and the HTT siRNA signals of all these plots are from the same samples tested in parallel with the candidate genes. The scrambled (n = 16) and HTT siRNA (n = 4) plots represent mean and SEM, whereas each of the two independently transfected wells of the hits are plotted individually. The genes targeted by siRNAs that obviously increased neuronal survival (higher survival at all the time points measured) were selected as potential hits.