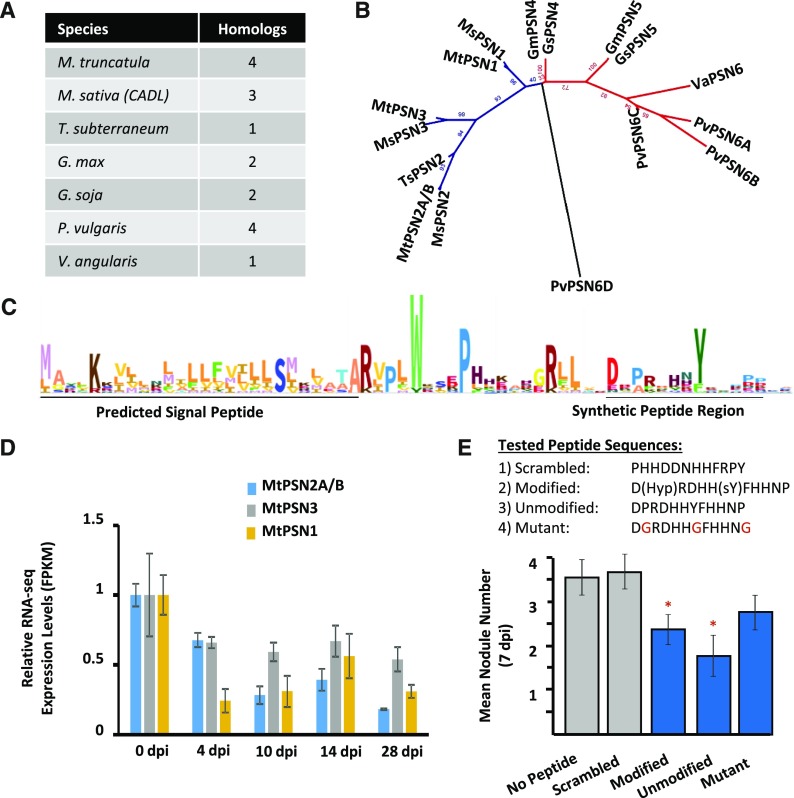
Figure 9.
A synthetic peptide from the novel PSN family inhibits nodulation. The novel PSN family was selected for testing based on expression patterns and legume-specific occurrence. Modified and unmodified versions of the predicted SSP from MtPSN1 were synthesized and tested for effects on nodulation, along with two control peptides. A, A total of 16 homologs of the PSN family were identified in seven legume species. The number of homolog sequences is indicated per species. B, Phylogenetic tree of the 16 homologs calculated by maximum likelihood using RAxML. Homologs separate into an IRLC (blue) and a bean clade (red). Species are indicated by two-letter code at the beginning of each label: Gm, G. max; Gs, G. soja; Ms, M. sativa; Mt, M. truncatula; Pv, P. vulgaris; Ts, T. subterraneum; Va, V. angularis. C, An HMM from the members of the PSN family is illustrated as a WebLogo. The N-terminal predicted signal peptide and the predicted bioactive peptide regions are indicated. D, Normalized RNA-seq expression levels (as FPKM) demonstrate the down-regulated expression of PSN family members in nodules. Each gene is normalized to the 0-dpi control. Error bars represent se (n = 3). E, Nodulation was induced with S. meliloti on agar plates embedded with the designated peptide or without peptide. Nodule numbers were counted 7 dpi. PTM-modified and -unmodified forms of the peptide, and to a lesser extent the conserved residue mutant peptide, suppressed nodule numbers, while the scrambled peptide did not affect nodule numbers. sY, Sulfated Tyr residue. Mutated residues in the mutant sequence are indicated in red. Error bars represent se (n = 20). Asterisks indicate statistically significant differences relative to no peptide control (*, P < 0.05).