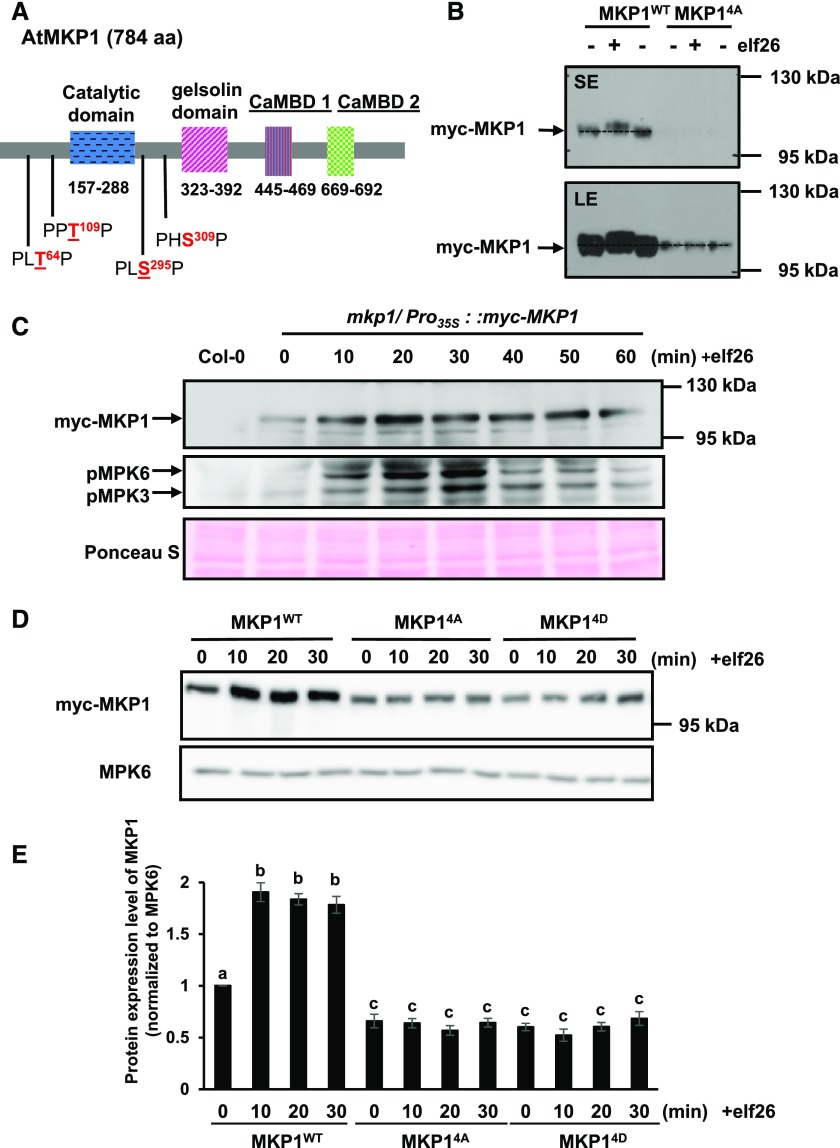
Figure 3.
Phosphorylation of MKP1 stabilizes the protein. A, Domain structure of the Arabidopsis MKP1 protein, showing the positions of the conserved putative MAPK phosphorylation sites. Underlined residues have been experimentally shown to be phosphorylated (see refs. in the text). B, Immunoblot analysis with anti-myc antibody of protein extracts from 14-d-old myc-tagged MKP1 (MKP1WT) or phosphorylation site mutant (MKP14A) transgenic seedlings treated with or without 1 µm elf26 for 20 min. Protein extracts were separated with 8% large-format (16 × 16 cm) SDS-PAGE. LE, Long exposure; SE, short exposure. C, Fourteen-day-old myc-tagged MKP1 transgenic seedlings were treated with 1 μm elf26 for the times indicated. Protein extracts from treated seedlings were separated by 8% mini-format (8.3 × 7.3 cm) SDS-PAGE and immunoblotted with anti-myc antibody to detect myc-MKP1 (top) or anti-phospho-p42/44 MAPK antibody to detect phosphorylated MAPKs (middle). Ponceau S staining of the membrane was used as a loading control (bottom). D, Fourteen-day-old transgenic seedlings expressing MKP1 wild-type protein (MKP1WT) or phosphorylation site substitutions (MKP14A and MKP14D) were treated with or without 1 µm elf26 for the times indicated. Protein extracts from treated seedlings were separated by 8% mini-format (8.3 × 7.3 cm) SDS-PAGE and immunoblotted with anti-myc antibody to detect the myc-MKP1 protein (top) or anti-MPK6 antibody as a loading control (bottom). This experiment was performed three times with similar results to those shown. E, Quantification of the western-blot band intensity of MKP1 protein normalized to the intensity of MKP6 used as a loading control. Graphed are means ± se, representative of three independent biological replicates (n = 3). Lowercase letters indicate significant groupings (P < 0.05). The statistical test was performed using ANOVA with Tukey’s pairwise comparison.