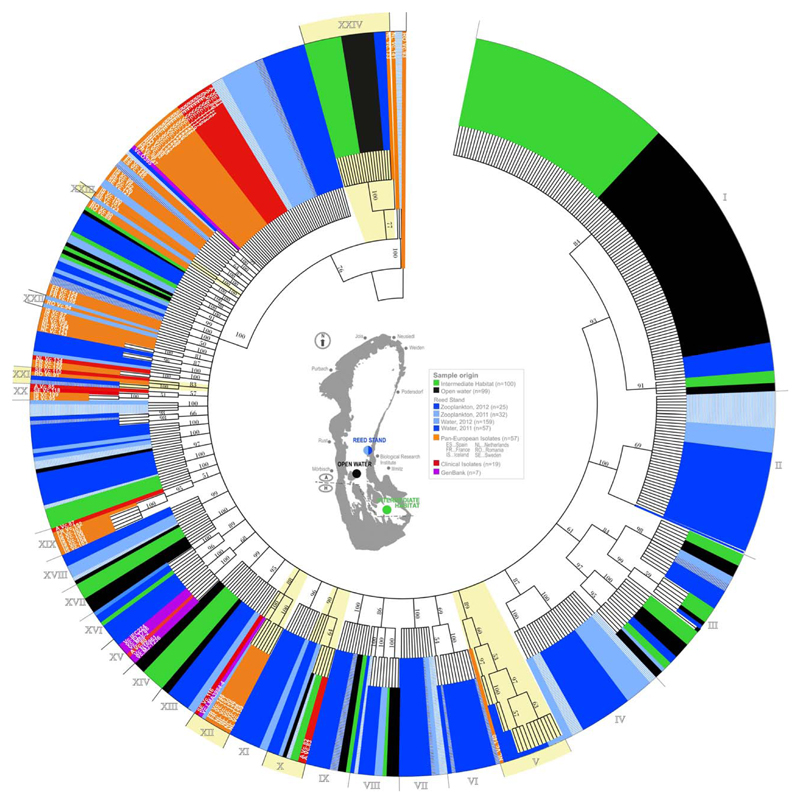
Fig. 2.
Maximum likelihood phylogenetic tree resulting from the analysis of the concatenated sequences comprising of gyrB, recA, pyrH and toxR (2270 bp) of 555 Vibrio cholerae isolates including 472 of lake Neusiedler See, 68 Pan-European isolates, 7 reference strains and 8 clinical isolates. Numerical values at nodes indicate bootstrap support (>50) obtained after 1000 replications. Clades discussed in text are given in Roman numerals. Colors denote origin of isolate detailed on the insert. Clades with mixed local and remote isolates (V, XII, XXI, XXIII and XXIV) and with mixed local environmental and clinical isolates (X) are marked with yellow. Insert depicts a schematic map of the lake, location of studied habitats and the sample type (water, zooplankton). Branch lengths are transformed to equal length for better visualization and do not represent true phylogenetic distances.