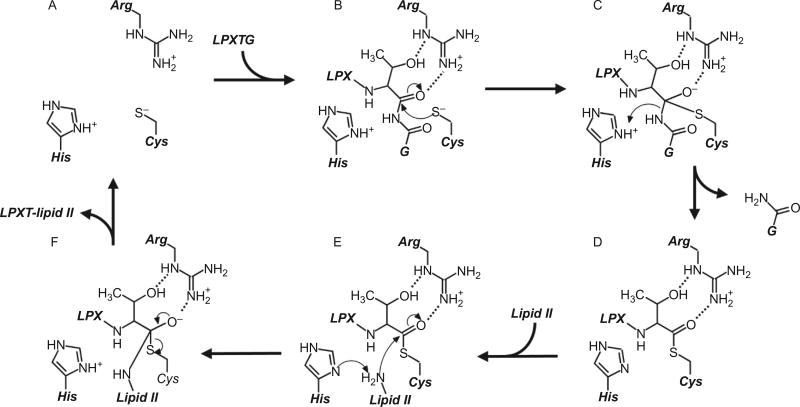
Fig. 7.
Molecular mechanism of sortase enzymes. The active site of sortase consists of a His–Cys–Arg triad, and in its active form, the His and Cys residues form a thiolate–imidazolium ion-pair (A). The reaction begins with recognition of an appropriate sorting signal (here, the LPXTG sorting signal for SrtA types is shown), and the active site Cys residue performs nucleophilic attack on the carbonyl carbon at the substrate’s P1 position (B). An oxyanion tetrahedral intermediate is stabilized by the nearby Arg residue that is likely oriented by interactions with the side chain of the substrate’s P1 residue, which is a threonine in over 95% of all substrates (C). The active His residue concomitantly donates a proton to the leaving group, and the tetrahedral transition state then collapses to form a semistable, thioacyl intermediate between the substrate’s P1 residue and the active site Cys (D). Next, the secondary substrate (here shown as lipid II used by cell wall anchoring sortases) enters the active site, where its terminal amine is deprotonated by the active His residue before performing nucleophilic attack on the carbonyl carbon in the thioacyl bond (E); this second tetrahedral intermediate (F) collapses to form a peptide bond between the two substrates, and the product is finally released to leave the regenerated active site (A).