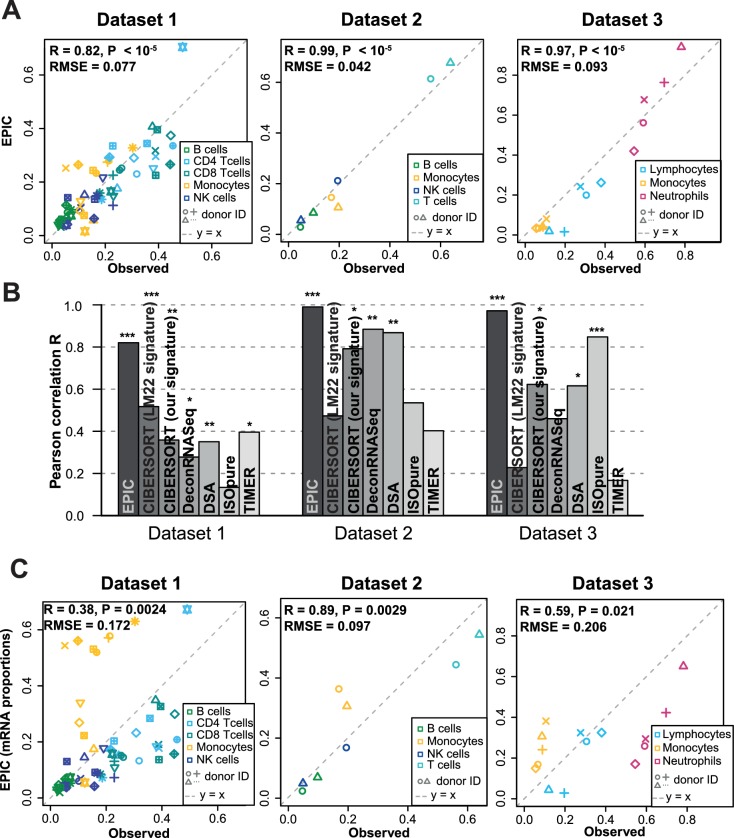
Figure 2. Predicting cell fractions in blood samples.
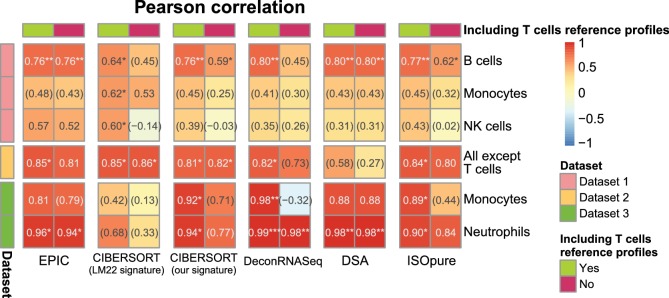
(A) Predicted vs. measured immune cell proportions in PBMC (dataset 1 (Zimmermann et al., 2016), dataset 2 (Hoek et al., 2015)) and whole blood (dataset 3 (Linsley et al., 2014)); predictions are based on the reference profiles from circulating immune cells. (B) Performance comparison with other methods. Significant correlations are indicated above each bar (*p<0.05; **p<0.01; ***p<0.001). (C) Predicted immune cells' mRNA proportions (i.e., without mRNA renormalization step) vs. measured values in the same datasets. Correlations are based on Pearson correlation; RMSE: root mean squared error. Proportions of cells observed experimentally are given in Supplementary file 3B-D.
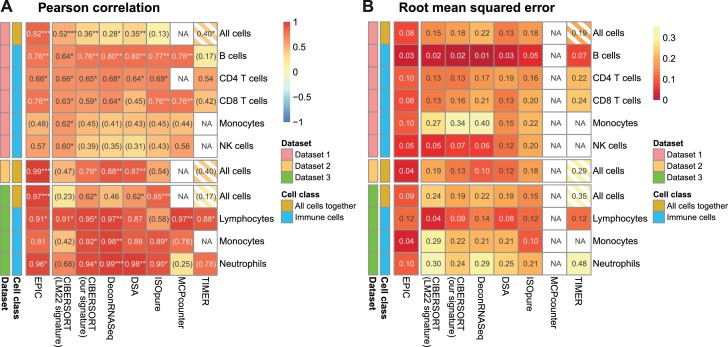
Figure 2—figure supplement 1. Comparison of multiple cell fraction prediction methods in blood datasets.
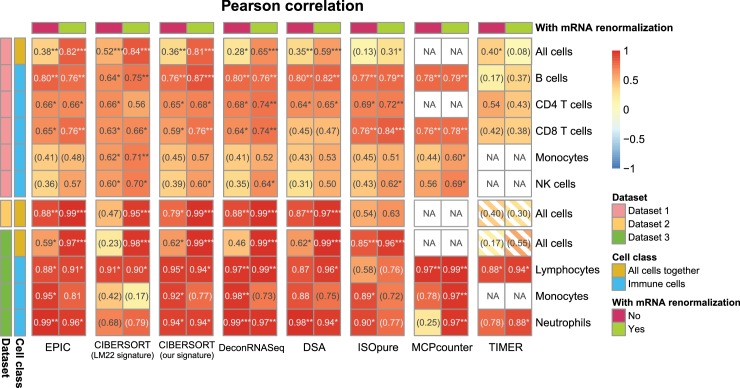
Figure 2—figure supplement 2. Effect of including an mRNA renormalization step for multiple cell fraction prediction methods.
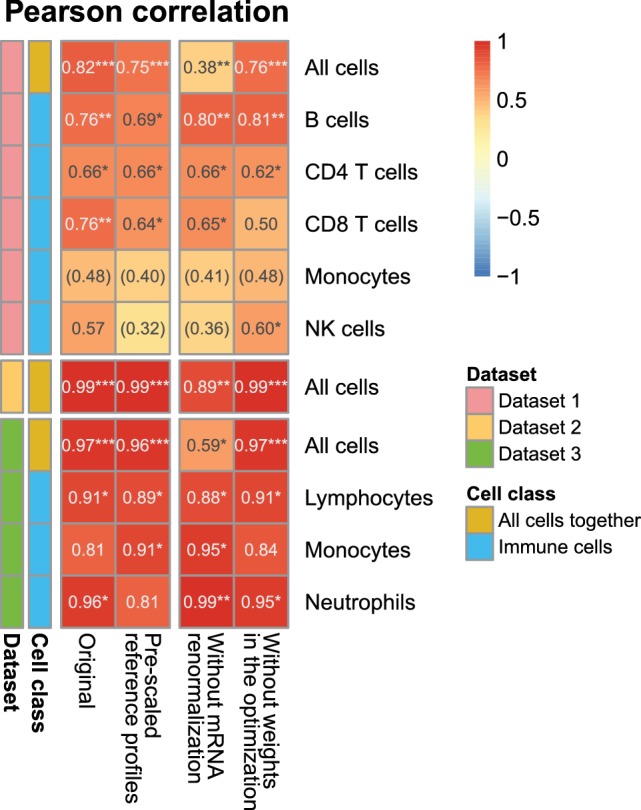
Figure 2—figure supplement 3. Effect of the various steps in EPIC on the prediction accuracy.