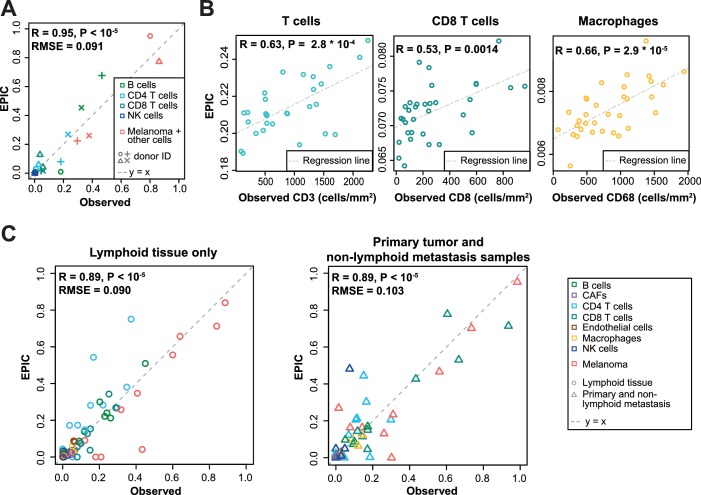
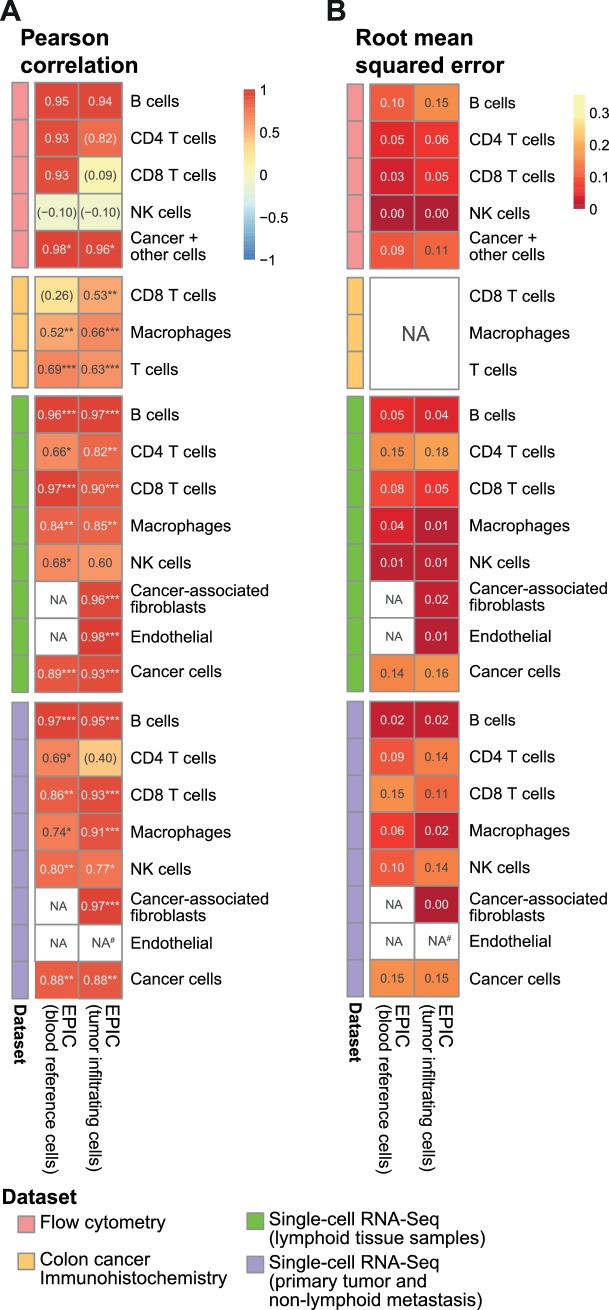
Figure 4. Predictions with reference profiles from tumor-infiltrating cells.
Same as Figure 3 but based on reference profiles built from the single-cell RNA-Seq data of primary tumor and non-lymphoid metastatic melanoma samples from Tirosh et al. (2016). (A) Comparison with flow cytometry data of lymph nodes from metastatic melanoma patients. (B) Comparison with IHC from colon cancer primary tumors (Becht et al., 2016). (C) Comparison with single-cell RNA-Seq data (Tirosh et al., 2016). For primary tumor and non-lymphoid metastasis samples, a leave-one-out procedure was used (see Materials and methods). Proportions of cells observed experimentally are given in Supplementary file 3A,E.