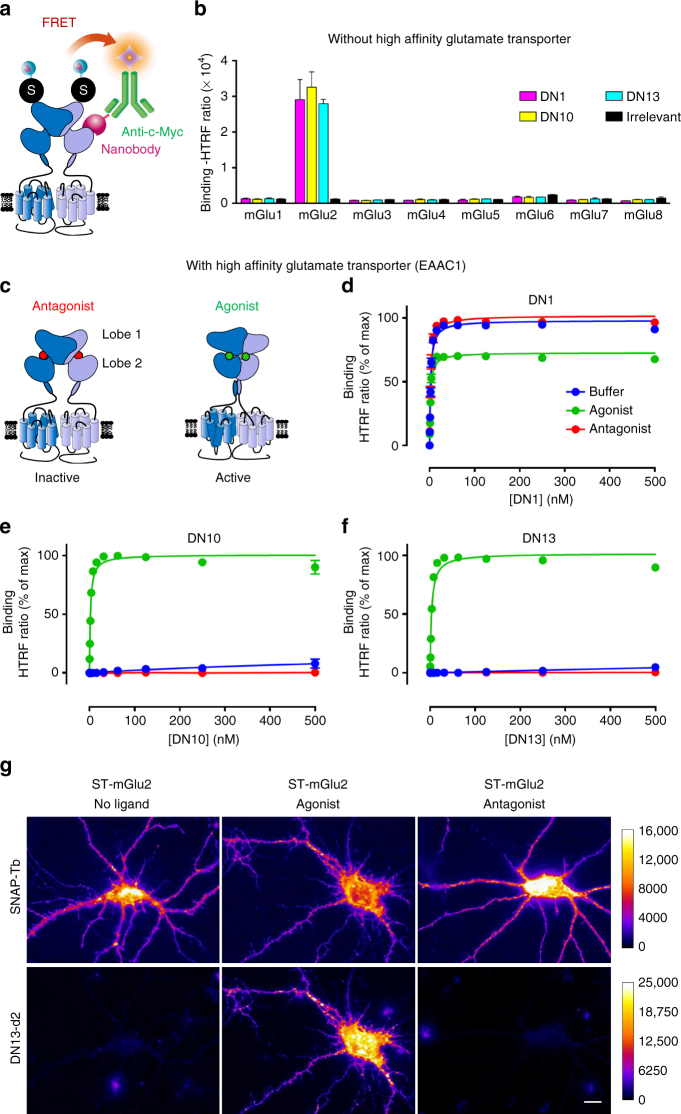
Fig. 1.
Nanobodies DN1, DN10, and DN13 specifically interact with mGlu2 receptors. a Cartoon illustrating the principle of the TR-FRET binding assay. The receptor fused to a SNAP-tag (black circled labeled “S”) is labeled with Lumi4-Tb (light blue ball) while the nanobody (purple) bearing a c-Myc epitope at its C-terminus is labeled with 200 nM of anti-c-Myc antibody (green) coupled to d2 fluorophores (orange). Binding of the nanobody to the receptor is then measured by a TR-FRET signal. b Specific TR-FRET binding data obtained with the indicated mGlu receptor and either DN1, DN10, DN13, or a control irrelevant nanobody in cells under basal conditions with ambient glutamate (no high affinity glutamate transporter being co-transfected). Data are mean ± SD of triplicates from a typical experiment representative of three experiments. c Cartoon illustrating the main active (right), and inactive (left) conformation of an mGlu2 dimer, stabilized by an agonist (glutamate or LY379268) or the antagonist LY341495, respectively. d–f Saturation binding curves obtained with DN1 d, DN10 e, and DN13 f on mGlu2 receptors under control conditions in cells co-expressing SNAP-mGlu2 and the high affinity glutamate transporter EAAC1 (buffer), in the presence of the agonist LY379268 (1 µM), or the antagonist LY341495 (10 µM). Data are mean ± SEM of three individual experiments each performed in triplicates. g Fluorescence signals on cultured hippocampal neurons transfected with SNAP-mGlu2 and labeled either with SNAP-Lumi4-Tb (top) or DN13-d2 (100 nM) (bottom), under basal, agonist (LY379268, 1 µM) or antagonist (LY341495, 10 µM) conditions. Scale bar: 20 µm