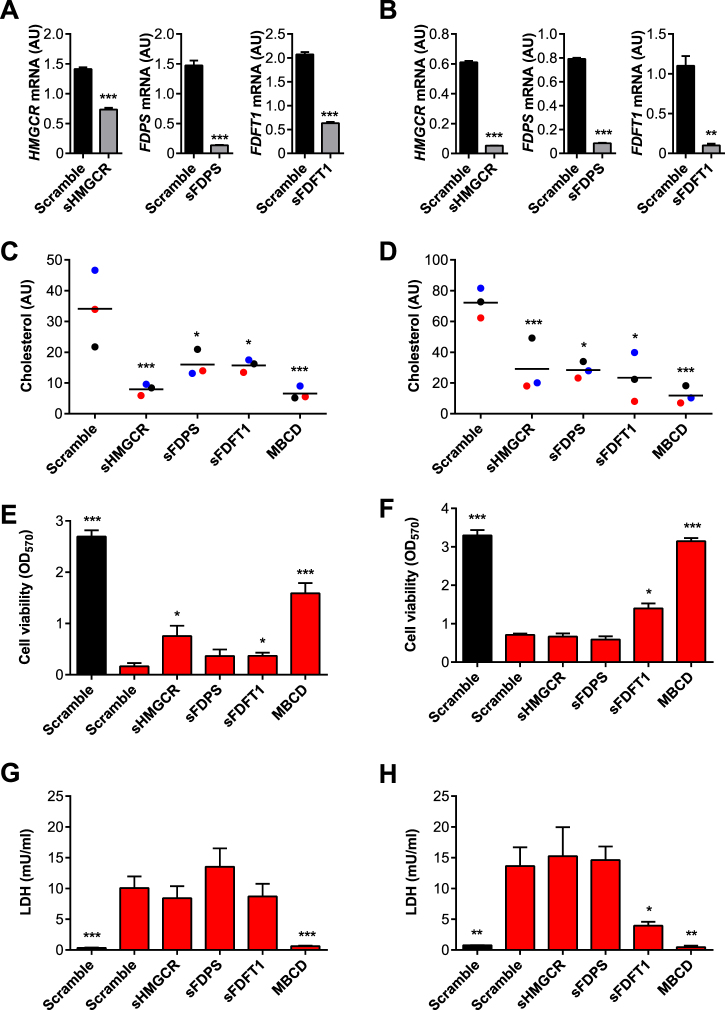
Figure 3.
RNA interference of mevalonate pathway enzymes. (A) BESC isolated from 3 animals, and (B) HESC from 3 independent passages, were transfected with scramble siRNA or siRNA targeting HMGCR, FDPS or FDFT1. The mRNA expression of each cognate gene was measured by qPCR, and data presented as mean (SEM) relative to two reference genes. Data were analyzed by Student’s t-test; values differ from scramble, **P < 0.01, ***P < 0.001. BESC isolated from 3 animals (C,E,G) or HESC from 3 independent passages (D,F,H) were incubated for 48 h in serum-free control medium, or media containing scramble siRNA or siRNA targeting HMGCR, FDPS or FDFT1, or cultured with methyl-β-cyclodextrin (MBCD) as a positive control. Cellular cholesterol (C,D) was measured and normalized to phospholipid concentrations, and data presented as arbitrary units (AU) with each dot representing an independent animal or cell passage, and a horizontal line indicating the mean. Cells were challenged with control media (■) or media containing PLO ( ) using 100 HU/ml for BESC and 200 HU/ml for HESC, for 2 h. Cell viability was quantified by MTT assay (E,F), and supernatants were collected to measure LDH (G,H). Data are presented as mean (SEM), and were analyzed by one-way ANOVA and Dunnett’s multiple comparison post-hoc test; values differ from scramble challenged with PLO,*P < 0.05, **P < 0.01, ***P < 0.001.
) using 100 HU/ml for BESC and 200 HU/ml for HESC, for 2 h. Cell viability was quantified by MTT assay (E,F), and supernatants were collected to measure LDH (G,H). Data are presented as mean (SEM), and were analyzed by one-way ANOVA and Dunnett’s multiple comparison post-hoc test; values differ from scramble challenged with PLO,*P < 0.05, **P < 0.01, ***P < 0.001.