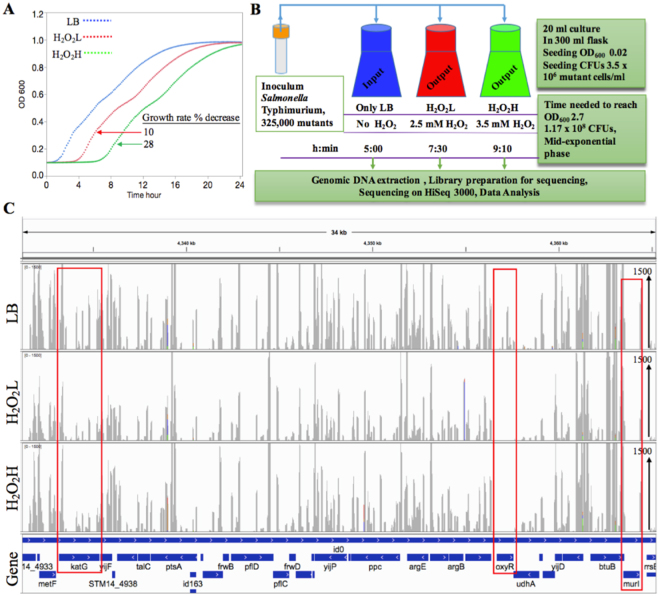
Figure 1.
Study design and identification of the genes required for H2O2 resistance using Tn-seq. (A) The effect of H2O2 on the growth rate of wild type S. Typhimurium. An overnight culture of bacteria was diluted 1:200 in the LB medium containing either 0 mM (control: blue), 2.5 mM H2O2 (H2O2L: red), or 3.5 mM H2O2 (H2O2H: green). The cultures were incubated at 37 °C for 24 h in a 96-well plate. The reduced growth rates in the presence of H2O2 were in reference to LB control. (B) Schematic representation of the Tn-seq experiment design. The Salmonella transposon mutant library was inoculated into LB, H2O2L and H2O2H. The three cultures were grown until they reached mid-exponential phase. Genomic DNA was extracted from each culture and subjected to library preparation, Illumina sequencing, and data analysis. (C) The Tn-seq profiles of the three conditions for the 34 kb genomic region starting from with metF gene and ending with rrsB. The vertical axis represents number of sequencing reads with the maximum of 1500 reads. The genes highlighted in red are katG, oxyR, and murI as examples of non-essential gene, conditionally essential gene, and essential gene, respectively.