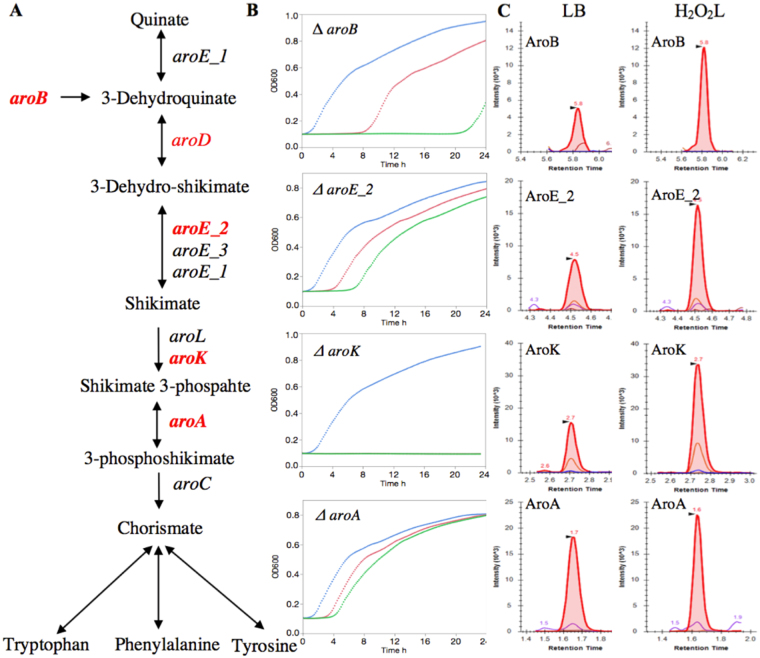
Figure 3.
The role of aromatic amino acid biosynthesis genes in resistance to H2O2. (A) Schematic representation of aromatic amino acid biosynthesis pathway, adapted from the KEGG pathway database. The genes in red color were identified by the Tn-seq analysis as conditionally essential for H2O2 resistance in Salmonella. The genes in bold type indicated that the mutant phenotype was also validated by individual mutant assay. (B) The overnight cultures of the individual mutants were diluted 1:200 in LB (no H2O2) and LB containing either 2.5 mM H2O2 (H2O2L) or 3.5 mM H2O2 (H2O2H). The cultures were incubated at 37 °C for 24 h in a 96-well plate. The growth curves are shown LB (blue), H2O2L (red), and H2O2H (green). In the growth curve of ΔaroK, the red color is overlapped with the green color. (C) Upregulation of Salmonella proteins in response to H2O2L in comparison to LB. Wild type S. Typhimurium strain was grown in LB, H2O2L, and H2O2H until mid-exponential phase. Targeted-proteomics was used to quantify AroB, AroE_2, AroK, and AroA protein expressions in response to H2O2L.