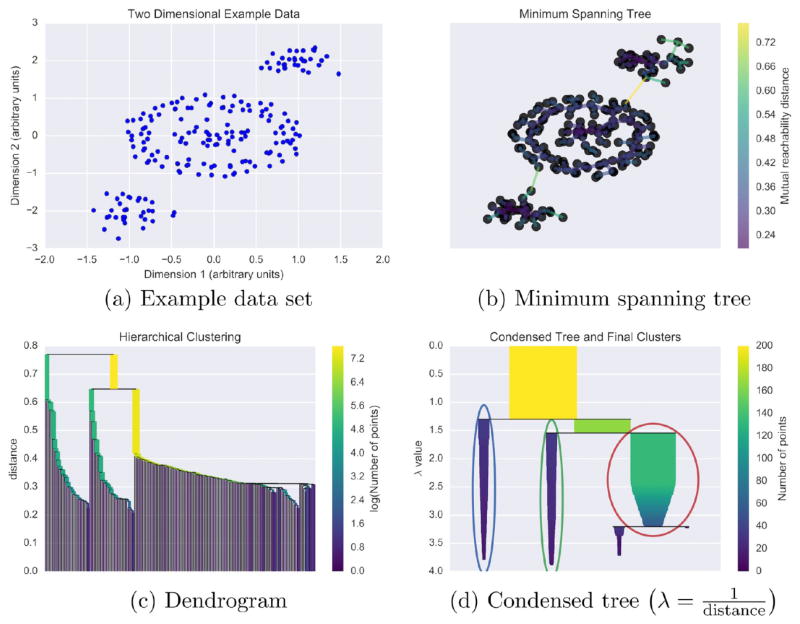
Figure 8.
Using a toy two-dimensional data set generated with scikit-learn140 (a), we give a conceptual explanation of HDBSCAN’s clustering algorithm. Using mutual reachability distance as the distance metric, a minimum spanning tree (b) is constructed. This tree solves an optimization problem such that removing any edge would create disconnected components–set(s) of nodes not connected to any other set(s). Based on the distances of connected points in the minimum spanning tree, single-linkage hierarchical clustering is performed (c). Splits in the dendrogram that create clusters smaller than the minimum cluster membership are rejected, and the final clusters (d) are determined using HDBSCAN’s novel cluster stability metric.35 In panel (d), λ is inverse distance. The example here is based on a tutorial by Leland McInnes, the author of the Python HDBSCAN implementation, available at github.com/lmcinnes/hdbscan. Additionally, we have made the Python code for reproducing our specific example available at figshare.com/articles/HDBSCAN_and_Amorim-Hennig_for_MD/3398266.