Abstract
HRAS, KRAS and NRAS, highly homologous proteins, are often mutationally activated in cancer. Usually, mutations cluster in codons 12, 13 and 61 and are detected by molecular genetic testing of tumor DNA. Recently, immunohistochemistry with SP174 antibody has been introduced to detect NRAS Q61R mutant protein. Studies on malignant melanomas showed that such an approach could be a viable alternative to molecular genetic testing. This investigation was undertaken to evaluate the value of SP174 immunohistochemistry for detection of NRAS Q61R mutant isoform. Two hundred ninety-two malignant melanomas were evaluated using Leica Bond-Max automated immunostainer. Twenty-nine tumors (10%) showed positive immunoreactivity. NRAS codon 61 was PCR amplified and sequenced in 24 positive and 92 negative cases using Sanger sequencing, qPCR and next generation sequencing approaches. A c.182A>G substitution leading to NRAS Q61R mutation was identified in 22 tumors. Two NRAS-wild type tumors revealed c.182A>G substitutions in H- and K-RAS codon 61, respectively. Both mutations were detected by next-generation sequencing and independently confirmed by Sanger sequencing. None of 85 NRAS codon 61-wild type tumors and 7 NRAS mutants other than Q61R showed immunoreactivity with SP174 antibody. Thus, SP174 antibody was 100% sensitive in detecting NRAS Q61R mutant isoform in malignant melanoma, but not fully specific as it cross-reacted with HRAS and KRAS Q61R mutant proteins. Therefore, molecular testing is needed to determine which RAS gene is mutated. The rarity of H-and K-RAS Q61R mutants in malignant melanoma let previous investigations erroneously conclude that SP174 is specific for NRAS Q61R mutant protein.
Keywords: RAS mutations, immunohistochemistry, H-, K-, and N-RAS Q61R-mutant proteins, SP174 antibody, Sanger sequencing, qPCR, Ion Torrent™ next-generation sequencing
INTRODUCTION
HRAS, KRAS and NRAS, highly homologous proteins, are located on the inner surface of the cell membrane. These enzymes represent GDP/GTP-regulated switches that transfer extracellular signals and play a fundamental role in signal transduction regulating cell proliferation, and apoptotic cell death.1
In cancer, RAS proteins are often pathologically activated. Typically, gain-of-function RAS mutations cluster in codons 12, 13 and 61. In Caucasian population approximately 20% of malignant melanomas carry NRAS mutations with codon 61 being the most commonly involved “hot spot”. Specifically, the c.182A>G substitution which leads to Q61R mutation accounts for about 40% of all NRAS mutants.2
For the last several decades, molecular genetic assays such as the melting curve analysis, Sanger sequencing, pyrosequencing and qPCR have been commonly used to detect RAS mutations. Lately, immunohistochemistry with SP174 antibody was introduced to pinpoint NRAS Q61R mutant protein, an equivalent of NRAS c.182A>G mutation. Recent studies on malignant melanomas showed that such immunohistochemical approach could be a valuable alternative to molecular genetic testing.3–9 This investigation was undertaken to verify value of SP174 immunohistochemistry in search for NRAS Q61R mutant protein in malignant melanomas.
MATERIAL and METHODS
Study material and design
Two hundred ninety-two anonymized, well characterized, malignant melanomas from Europe and the United States were analyzed. Malignant melanoma diagnosis was based on clinicopathologic data, histology and S100, HMB45 and Melan A immunohistochemistry. Clinicopathologic characteristic of analyzed cases is summarized in Table 1. The cohort included 96 primary tumors, 187 metastatic lesions and 9 cases in which primary tumor versus metastatic status was unclear. SP174 immunohistochemistry was performed in the Laboratory of Pathology on multitissue blocks build as previously reported.10 DNA was extracted from formalin-fixed paraffin-embedded tumor tissues following published procedure.11 Screening for RAS mutations was completed independently and blindly without knowledge of the results from immunohistochemical studies in three different institutions. Sanger sequencing in the Laboratory of Pathology, qPCR in the Department of Biology and Genetics, University of Gdansk, Gdansk, Poland and Ion Torrent™ next-generation sequencing in the Department of Molecular Diagnostics, Holycross Cancer Center, Kielce, Poland.
Table 1.
Summary of clinicopathologic data.
| Age* | 18 to 96 (median 62) |
|---|---|
| Sex** | |
| Female | 127 |
| Male | 109 |
| Primary tumors | 96 |
| Acral | 2 |
| Anorectal | 38 |
| Esophageal | 9 |
| Eyeball | 2 |
| Nasal mucosa | 1 |
| Skin | 17 |
| Metastatic tumors | 187 |
| Adrenal | 1 |
| Bone | 2 |
| Brain | 6 |
| Intestine | 14 |
| Kidney | 3 |
| Liver | 11 |
| Lymph node | 57 |
| Lung | 8 |
| Pelvis | 2 |
| Retroperitoneum | 1 |
| Skin | 33 |
| Soft tissue | 27 |
| Unknown | 22 |
| Unknown | 9 |
Age based on 228 cases;
Sex o based on 236 cases
Immunohistochemistry
A rabbit monoclonal antibody, clone SP174 (Spring™ Bioscience, Pleasanton, CA) and Leica Bond-Max automatic immunostainer (Leica, Bannockburn, IL) with Leica Refine detection kit were used in this study. The primary antibody was incubated for 30 min, followed epitope retrieval using Leica H2 buffer (25 min). The 1:100 dilution of primary antibody was selected as the lowest dilution yielding a strong signal in NRAS Q61R mutant while giving no staining in NRAS wild type tumor. Either Diaminobenzidine or Fast Red were used for visualization following protocols provided by Leica. The immunostainings were scored arbitrarily by three pathologists (S.I., J.L., M.M.) as negative (no staining), weakly positive and strongly positive.
Molecular studies
HRAS, KRAS and NRAS codon 61 sequences were PCR amplified and the amplification products were evaluated by Sanger sequencing as previously reported.11 Primer sequences and PCR conditions for each reaction are listed in Table S1 in supplementary data. NRAS and KRAS qPCR assays were performed using RAS Mutation Analysis Kits (NRAS- and KRAS-RT50) and Rotor-Gene Q Software version 1.7 following the manufacturer’s instructions (EntroGen, Inc., Woodland Hills, CA). These tests detect spectrum of NRAS and KRAS substitutions leading to NRAS Q61H, -K, -L, and -R, and KRAS Q61H-L, and R mutations at the protein level. Next-generation sequencing was completed using the Ion Torrent™ next-generation sequencing platform and Ion AmpliSeq™ Cancer Hotspot Panel v2 Kit following the manufacturer’s instructions (Life Technologies/Thermo Fisher Scientific, Waltham, MA) and previously published procedure.12
RESULTS
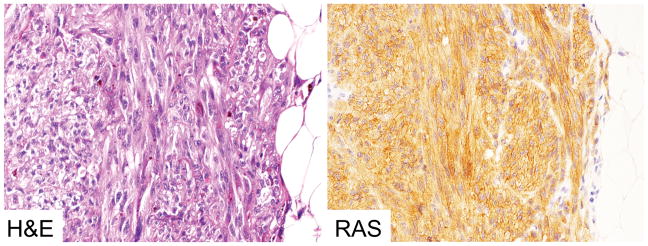
Twenty-nine (10%) of 292 analyzed malignant melanomas revealed positive immunostaining with SP174 antibody. In all cases, delicate granular perimembranous and cytoplasmic staining was seen. Example of SP174 immunostaing is shown in Figure 1.
Figure 1.
Example of positive immunoreactivity to SP174, NRAS Q61R mutant-specific antibody in malignant melanoma.
Twenty-four SP174-positive cases with tissue available for DNA extraction were evaluated by Sanger sequencing. A c.182A>G substitution leading to NRAS Q61R mutation at the protein level, was identified in 11 of 24 analyzed cases. In the remaining SP174 positive cases: 1) PCR amplification was unsuccessful (n=4), 2) Sanger sequencing chromatograms showed a small mutant peak and the possibility of c.182A>G substitution could not be ruled out (n=7), 3) only wild type NRAS codon 61 sequences were detected (n=2). Subsequently, these cases (n=13) were blindly evaluated using qPCR and Ion Torrent™ next-generation sequencing. NRAS codon 61 mutants were identified in all but 2 tumors. The latter revealed c.182A>G substitution in H-and K-RAS codon 61 (Q61R mutation), respectively. The next-generation sequencing results were later confirmed by PCR amplification of HRAS and KRAS codon 61and Sanger sequencing.
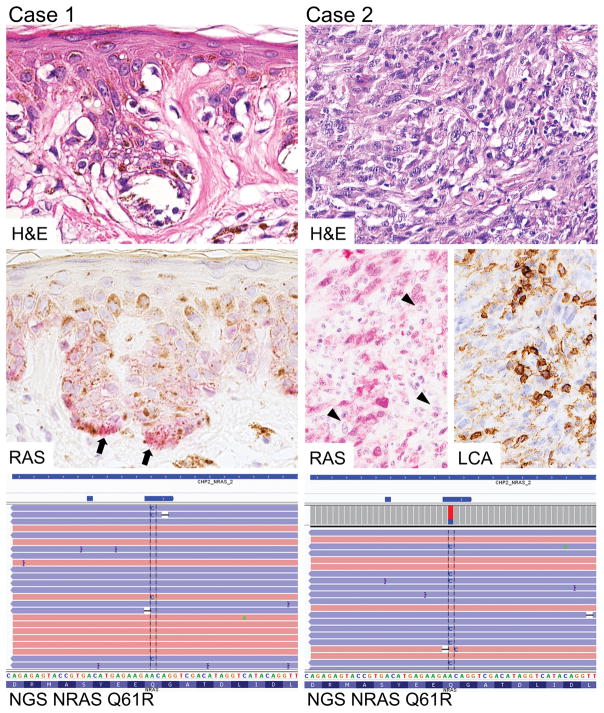
In 3 cases, Ion Torrent™ next-generation sequencing detected c.182A>G NRAS substitutions with moderate to low (35%, 27% and 6%) frequency. In these cases, the material submitted for Sanger sequencing was re-examined and either inadequate sampling, dominance of normal stromal cells and lymphoid elements, or low tumor cell content was identified to explain negative results in mutation analysis. Representative examples of histology, immunohistochemistry and next-generation sequencing are shown in Figure 2. Representative examples of histology, immunohistochemistry and molecular genetic testing are shown in Figure 3.
Figure 2.
Two malignant melanomas, one with low tumor cell content, second with prominent stromal cells and lymphoid elements (LCA staining). Next generation sequencing revealing NRAS c.182A>G substitution predicted to cause NRAS Q61R mutation.
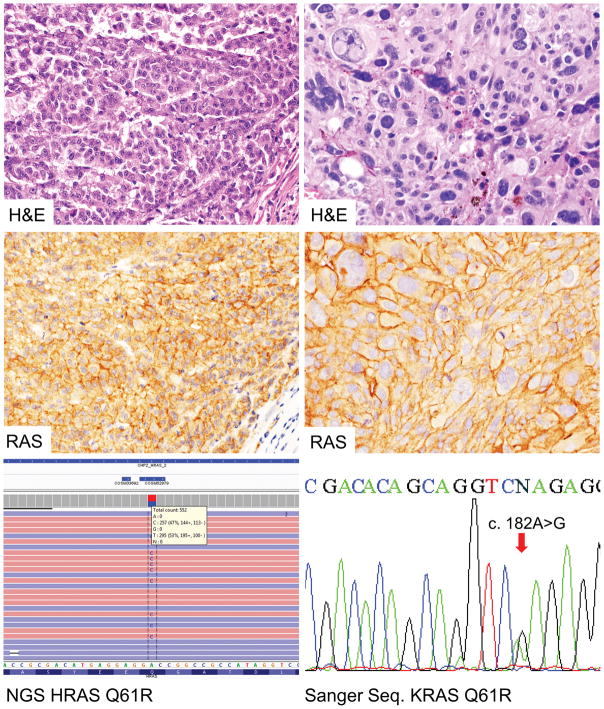
Figure 3.
Two malignant melanomas with positive immunoreactivity to SP174 (wild type for NRAS codon 61 by Sanger sequencing) carry HRAS or KRAS c.182A>G substitutions documented by next generation sequencing and sanger sequencing, respectively.
None of 85 NRAS codon 61-wild type tumors and 7 NRAS mutants other than Q61R showed immunoreactivity with SP174 antibody. However, weak SP174 immunostaining was noticed in the normal sebaceous glands in a few cases. Representative images are shown in Figure S1 in supplementary data.
DISCUSSION
HRAS, KRAS and NRAS proto-oncogenes are often mutated in cancer including malignant melanoma. The frequency of the mutant isoforms defined by distinctive point mutations in RAS codons 12, 13 and 61 varies significantly between different types of tumors.2 In malignant melanoma, NRAS Q61R is the most common RAS mutation while KRAS Q61R is extremely rare as reported by the catalog of somatic mutations in cancer (cancer.sanger.ac.uk/cosmic). However, HRAS Q61-mutants are distinctive feature of Spitz tumors with benign or unknown malignant potential.13
In vitro studies based on colorectal cancer cell lines showed that oncogenic potential and sensitivity to inhibitor treatment can depend on type of RAS mutant isoform expression. Thus, in the future, RAS mutation status might be a key element necessary for pinpointing right therapeutic strategy in malignant melanoma.14
Recently published investigation showed that immunohistochemistry with SP174, a rabbit monoclonal antibody, detects NRAS Q61R mutant protein with high sensitivity and specificity, and because of that it may have clinical utility and substitute molecular genetic testing.3–9 Although this study confirmed high (100%) sensitivity of SP174 immunohistochemistry in detection of NRAS Q61R mutant isoform, specificity of this antibody has to be questioned because of cross-reactivity with HRAS- and KRAS-Q61R mutant proteins. This cross-reaction can be attributed to the high sequence homology of the amino-terminal catalytic domain (amino acid 1–165) of RAS proteins.15 Previously emphasized 100% specificity of this antibody was based on the presumption rather than experimental evidence because no malignant melanoma HRAS or KRAS Q61R mutants were evaluated.5 During the preparation of this manuscript cross-reactivity of SP174 with H-, and K-RAS Q61R mutants has been reported in colorectal and thyroid cancer.16,17
Weak non-specific staining of normal cells including adipocytes, bronchial epithelial cells, endothelial cells macrophages and plasma cells with SP174 antibody was reported in previously published studies.4,6,8 In this study, only normal sebaceous glands showed weak SP174 positivity in a few analyzed cases. Such cross-reactivity may be attributed to the technical factors including platform used for the immunohistochemistry. A cross-reactivity between mutant-specific antibody and normal cell epitopes was previously reported for VE1, BRAF V600E mutant-specific antibody.11,18
For the last several decades Sanger sequencing has been considered the gold standard for DNA mutation detection. However, sensitivity of this methodology doesn’t allow discovery of the mutations in the tissue samples with low tumor cell content. Although laser-based micro dissection could be employed to enriched tumor cell population, the procedure is time consuming and can’t be easily used in clinical setting. Next-generation sequencing used in this study is a powerful tool allowing detection of the low-copy mutant alleles in partially degraded DNA from formalin-fixed-paraffin-embedded tissue.19
This study showed that prescreening with SP174 immunohistochemistry followed by next-generation sequencing offers effective and precise detection of different RAS codon 61 mutants. Significant cost reduction could be an additional advantage of such strategy. Although availability of next-generation sequencing technology is still restricted to research/academic centers, recently developed smaller, less expensive instruments with smaller targeted mutation panels may be more applicable for clinical testing.20
This study showed that immunohistochemistry with SP174 antibody allowed identification of three different H-, K- and N-RAS Q61R mutant proteins. Although this antibody is 100% sensitive for RAS Q61R, molecular genetic testing is necessary to determine which RAS gene is mutated.
Supplementary Material
References
- 1.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 2.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy MJ, Elaba Z. Potential utility of mutant oncogene-specific antibodies in melanoma. Am J Dermatopathol. 2014;36:522–523. doi: 10.1097/DAD.0b013e318292b396. [DOI] [PubMed] [Google Scholar]
- 4.Ilie M, Long-Mira E, Funck-Brentano E, et al. Immunohistochemistry as a potential tool for routine detection of the NRAS Q61R mutation in patients with metastatic melanoma. J Am Acad Dermatol. 2015;72:786–793. doi: 10.1016/j.jaad.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Massi D, Simi L, Sensi E, et al. Immunohistochemistry is highly sensitive and specific for the detection of NRASQ61R mutation in melanoma. Mod Pathol. 2015;28:487–497. doi: 10.1038/modpathol.2014.137. [DOI] [PubMed] [Google Scholar]
- 6.Uguen A, Talagas M, Costa S, et al. NRAS (Q61R), BRAF (V600E) immunohistochemistry: a concomitant tool for mutation screening in melanomas. Diagn Pathol. 2015;10:121. doi: 10.1186/s13000-015-0359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dias-Santagata D, Su Y, Hoang MP. Immunohistochemical Detection of NRASQ61R Mutation in Diverse Tumor Types. Am J Clin Pathol. 2016;145:29–34. doi: 10.1093/ajcp/aqv015. [DOI] [PubMed] [Google Scholar]
- 8.Just PA, Pouliquen C, Audebourg A, et al. High specificity and sensitivity of NRAS Q61R immunohistochemistry (IHC) in melanomas. J Am Acad Dermatol. 2016;74:572–573. doi: 10.1016/j.jaad.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Kakavand H, Walker E, Lum T, et al. BRAFV600E and NRASQ61L/Q61R mutation analysis in metastatic melanoma using immunohistochemistry: a study of 754 cases highlighting potential pitfalls and guidelines for interpretation and reporting. Histopathology. 2016;69:680–686. doi: 10.1111/his.12992. [DOI] [PubMed] [Google Scholar]
- 10.Miettinen M. A simple method for generating multitissue blocks without special equipment. Appl Immunohistochem Mol Morphol. 2012;20:410–412. doi: 10.1097/PAI.0b013e318245c82f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasota J, Kowalik A, Wasag B, et al. Detection of the BRAF V600E mutation in colon carcinoma: critical evaluation of the imunohistochemical approach. Am J Surg Pathol. 2014;38:1235–1241. doi: 10.1097/PAS.0000000000000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopczynski J, Kowalik A, Chłopek M, et al. Oncogenic Activation of the Wnt/β-Catenin Signaling Pathway in Signet Ring Stromal Cell Tumor of the Ovary. Appl Immunohistochem Mol Morphol. 2016;24:e28–33. doi: 10.1097/PAI.0000000000000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Engen-van Grunsven AC, van Dijk MC, Ruiter DJ, et al. HRAS-mutated Spitz tumors: A subtype of Spitz tumors with distinct features. Am J Surg Pathol. 2010;34:1436–1441. doi: 10.1097/PAS.0b013e3181f0a749. [DOI] [PubMed] [Google Scholar]
- 14.Haigis KM, Kendall KR, Wang Y, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 16.Turchini J, Andrici J, Sioson L, et al. NRASQ61R Mutation-specific Immunohistochemistry is Highly Specific for Either NRASQ61R or KRASQ61R Mutation in Colorectal Carcinoma. Appl Immunohistochem Mol Morphol. 2016 Feb 9; doi: 10.1097/PAI.0000000000000333. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 17.Reagh J, Bullock M, Andrici J, et al. NRASQ61R Mutation-specific Immunohistochemistry Also Identifies the HRASQ61R Mutation in Medullary Thyroid Cancer and May Have a Role in Triaging Genetic Testing for MEN2. Am J Surg Pathol. 2016 Sep 15; doi: 10.1097/PAS.0000000000000740. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 18.Jones RT, Abedalthagafi MS, Brahmandam M, et al. Cross-reactivity of the BRAF VE1 antibody with epitopes in axonemal dyneins leads to staining of cilia. Mod Pathol. 2015;28:596–606. doi: 10.1038/modpathol.2014.150. [DOI] [PubMed] [Google Scholar]
- 19.Koboldt DC, Steinberg KM, Larson DE, et al. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.