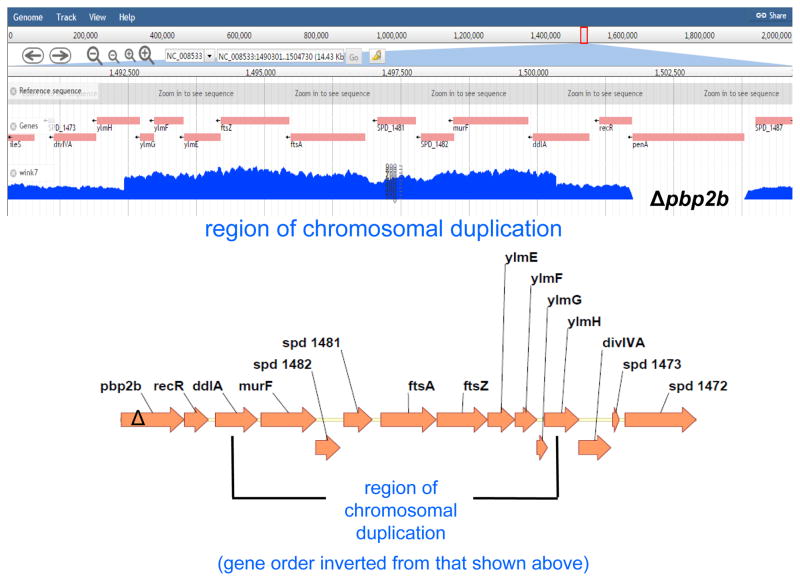
Fig. 6.
The chromosomal region surrounding ftsA-ftsZ (ddl′-ylmH′) is tandemly duplicated in the Δpbp2b sup6 suppressor strain. Top, number of reads from Illumina NextSeq DNA sequencing showing that the ddl′-ylm′ region is duplicated in the Δpbp2b sup6 suppressor strain (Table S2). No other mutations were detected in this strain. The number of reads across the duplicated region is ≈twice that of the adjoining regions. As expected, there are no reads from penA (pbp2b), which is deleted in the Δpbp2b suppressor strain. Reads spanning the junction between the 3′ end of the repeat region to the 5′ end of the repeat region were identified by bioinformatic analysis described in Experimental procedures, indicating that the repeat is a single tandem duplication that occurred in place without any other obvious re-arrangements taking place. Bottom, diagram of the annotated chromosomal region of the Ω(ddl′-ylm′) region, including the ftsA-ftsZ operon, duplicated in the Δpbp2b sup6 suppressor.