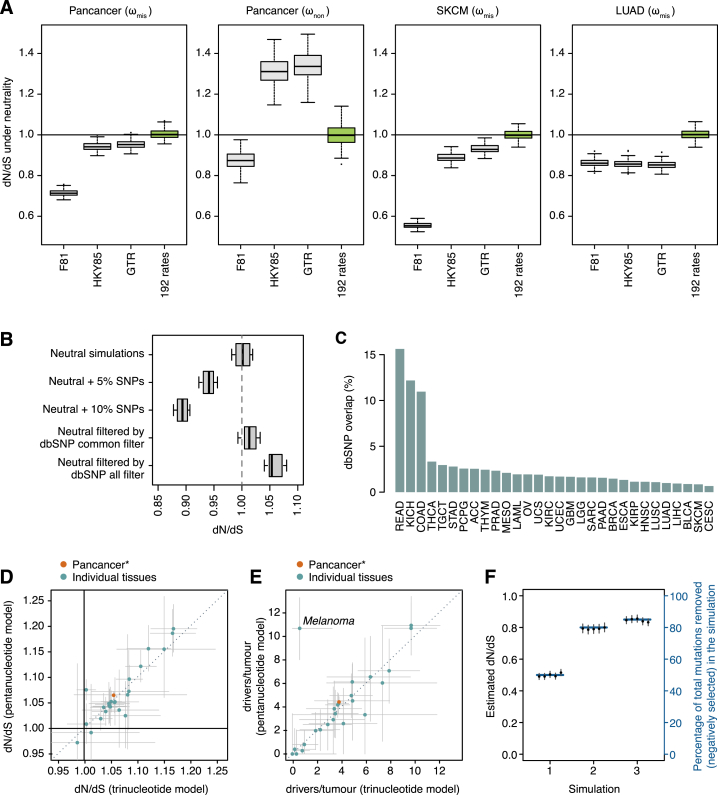
Figure S1.
Impact of Different Confounding Factors on Analyses of Selection, Related to Figures 1–5
This includes simplistic substitution models, SNP contamination, SNP filtering and inadequate background models of the variation of the mutation rate.
(A) Impact of simplistic mutation models on the accuracy of dN/dS in different scenarios. Each boxplot represents the dN/dS ratios estimated from 100 neutral simulations of 10,000 random coding substitutions. To exemplify the impact on dN/dS of different mutational spectra, we simulated neutral datasets using the trinucleotide spectra observed in the three different cohorts of samples (pancancer, melanoma and lung adenocarcinoma). Different panels depict dN/dS ratios for missense (ωmis) or nonsense (ωnon) mutations.
(B) Simulations of the impact on dN/dS of germline SNP contamination and SNP over-filtering in catalogs of somatic mutations. 10 neutral datasets were generated by local randomization of 607 cancer whole-genomes (Alexandrov et al., 2013). Datasets with varying degrees of germline SNP contamination were simulated by adding 5% or 10% of germline common SNPs (minor allele frequency > = 5%) from 1000 genomes phase 3 (Auton et al., 2015) to the neutral simulations. Datasets with varying levels of SNP over-filtering were simulated by removing any mutation from the neutral datasets that overlapped a polymorphic site in dbSNP build 146 (either using common sites or all sites) (Sherry et al., 2001).
(C) Percentage of mutations from the public TCGA catalogs of somatic calls that overlap a common dbSNP site. Based on simulations, an overlap of 1%–3% might be expected depending on the dominant mutational signatures present in a dataset, but several public TCGA catalogs show a much higher overlap suggesting extensive germline SNP contamination. As predicted from (B), this leads to an artifactual signal of negative selection in these datasets (STAR Methods).
(D) Consistency between genome-wide dN/dS estimates using the trinucleotide and pentanucleotide substitution models across cancer types. Green dots represent genome-wide dN/dS estimates for each cancer type separately, and the orange dot depicts the pancancer estimates (using the 24 cancer types with CaVEMan mutation calls).
(E) Corresponding estimates of the average number of driver coding substitutions per tumor. For the purpose of estimating the excess of mutations from dN/dS ratios, dN/dS values below 1 are set to 1. Error bars depict 95% CIs.
(F) Simulations demonstrating the validity of estimating dN/dS at a cohort level, in heterogeneous cohorts of samples without patient-specific substitution models. The three scenarios simulated include extreme examples of heterogeneous mixtures of samples with variable signatures, numbers of mutations and selection. In each scenario, the correct fraction of mutations removed by negative selection across samples is shown as a blue horizontal line (right y axis). Estimated dN/dS values from five simulations of each scenario are shown as dots with CIs (left y axis).