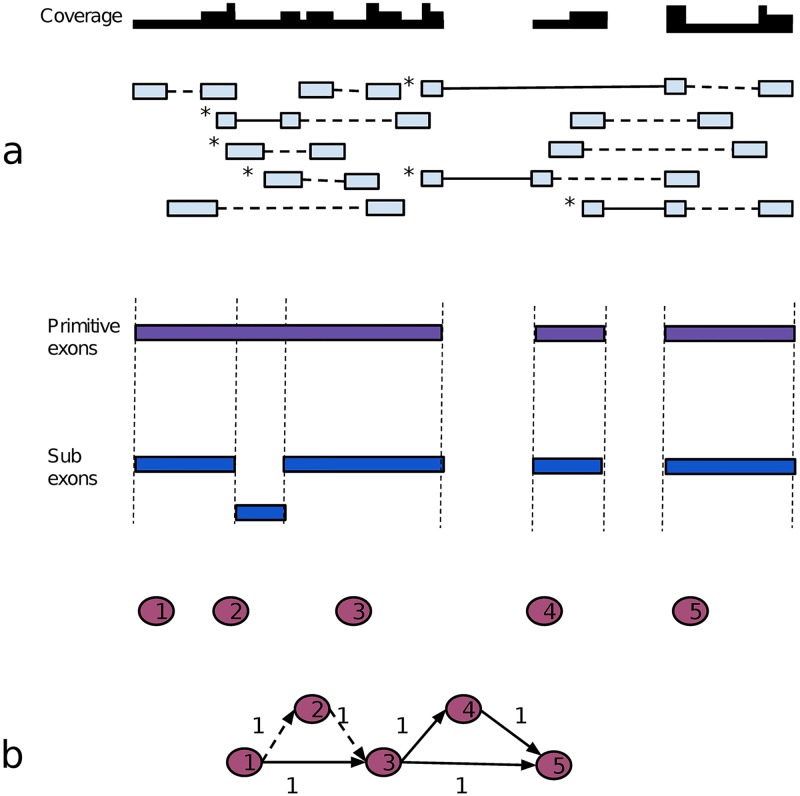
Fig 8. Translation of read alignments into a splicing graph.
(a) Eleven imaginary aligned paired-end reads (or read-pairs) are represented by light blue boxes intersected by solid lines, which indicate splicing junctions, and broken lines, which indicates gap sequences. Above the read-pairs, the coverage plot is shown. The white regions have zero coverage. Below the read-pairs, three primitive exons are shown as purple boxes and five subexons in dark blue, numbered from 1–5. (b) The splicing graph constructed from part (a). The numbered nodes in the splicing graph are subexons from part (a). Dashed Arrows represent the non-intron edges and solid arrows indicate the intron edges. The numbers next to edges are the weights(number of read-pairs supports). A read-pair that contributes to an edge weight is stressed using an asterisk near its upper-left corner. All the arrows also indicate the transcription direction. The source node and target node in the splicing graph are not shown.