Figure 1.
Virus genome data from EBOV cases in Sierra Leone characterizes the spatial spread of the epidemic. (a) The time course is shown for the number of confirmed cases [40] and sequenced EBOV genomes [33]. Three stages of the epidemic are highlighted. (b) Genetic linkages are illustrated with ancestors (open circles) and descendants (closed dots), both coloured by the origin district shown in the map key. The blue arrow highlights a linkage from the Western Urban to Kenema districts. (c) Chiefdom populations (greyscale) and major roads (yellow traces) are illustrated on the map of Sierra Leone. The blue arrow highlights the fastest driving route between the Western Urban to Kenema district. (d) All transmission distances are shown in a cCDF. The distribution of transmission distances are fit by a power law with ρ = 1.66. The blue arrow follows the linkage from (b) and (c). (e) Two spatial models are plotted as maps representing the probability of observing a new case linked to the Western Urban district, using (ρ⋆ = 1, τ⋆2 = 1) for the gravity model and ρ = 1.66 for the power law. (f) The log-likelihood ratio, 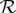 , comparing the gravity and power-law models, is plotted for 50-day windows. The dashed black line represents (ρ = 1.66, τ2 = 1) fixed in time; the solid black line of
, comparing the gravity and power-law models, is plotted for 50-day windows. The dashed black line represents (ρ = 1.66, τ2 = 1) fixed in time; the solid black line of 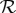 uses the MLE (ρ⋆(t), τ⋆2(t)), computed for each window. The solid red trace describes the number of linkages.
uses the MLE (ρ⋆(t), τ⋆2(t)), computed for each window. The solid red trace describes the number of linkages.

