Fig. 7.
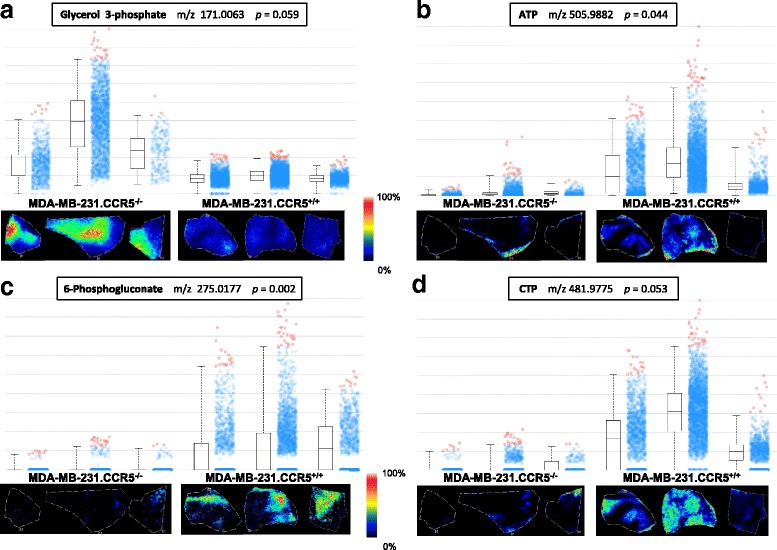
MALDI-FTICR-MSI analysis confirms that MDA-MB-231.CCR5−/− cells are less metabolically active than MDA-MB-231.CCR5+/+ cells. Tumor tissue was harvested at tumor onset from NSG mice that had received MDA-MB-231.CCR5−/− or MDA-MB-231.CCR5+/+ tumor cell suspensions, rapidly frozen, then sectioned in a cryostat, sprayed with 9AA matrix and subjected to MSI using MALDI-FTICR and a raster width of 50 μm over a m/z range of 150-1100. Each panel (a-d) represents an identified metabolite. Each box-plot contains a rectangle divided by a horizontal line, which represents the median intensity of that metabolite. Lower and upper bounds of the box represent the second and third quartile. Lines extending vertically from the box represent lower and upper quartiles (0% and 99% respectively). The cloud part of the plot shows how spectra from a given region are spread by intensity for each metabolite and region. Blue dots represent the spectra in-which intensities are between the lower and upper quartiles. Red dots represent outliers. Images below the box plots are show the relative abundance of each metabolite for each tissue (normalized to TIC) and based on the intensity scale provided
