Figure 3.
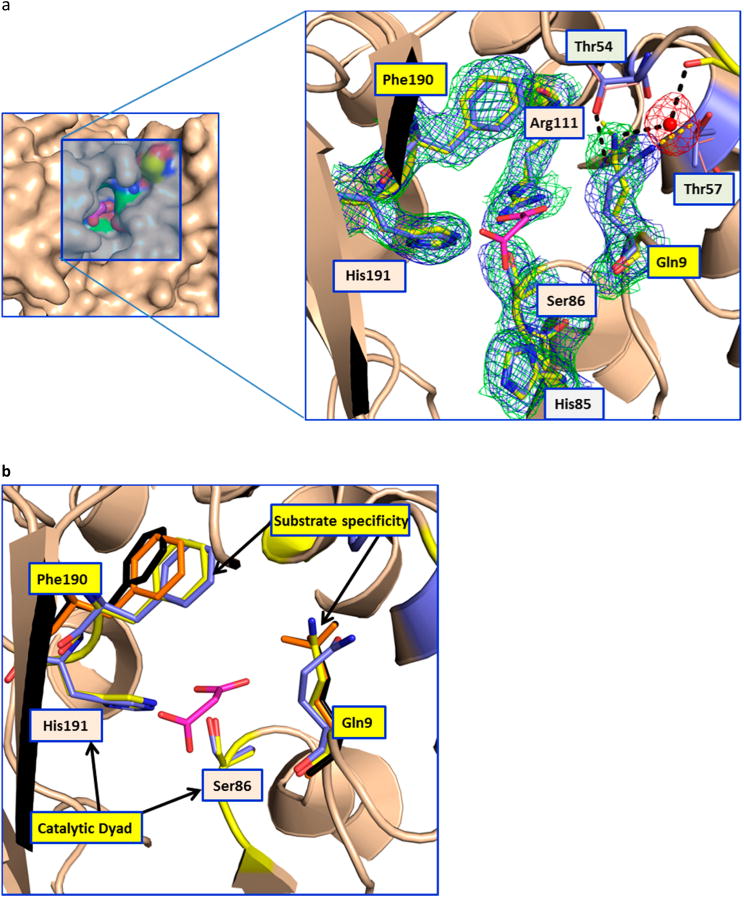
Conformational changes in the AT. (a) The left panel shows the surface representation of the AT active site (apo-LCLS structure), highlighting its deep narrow pocket. The close-up view of the malonate binding site is shown at the right. In the close-up view, the catalytic dyad residues (Ser86 and His191) are labeled with orange backgrounds and the gatekeeper residues (Gln9 and Phe190) are labeled with yellow backgrounds. Thr54 and Thr57, which are involved in the stabilizing Gln9, are also shown. The hydrogen bonds in the room-temperature and 100 K structures are shown as black and yellow dashed lines, respectively. Gln9 in the room-temperature structure is stabilized by a water molecule that bridges to the main chain carbonyl of Ser52. The 2Fo – Fc maps contoured at 1σ for the room-temperature and 100 K structures are shown as green and blue meshes, respectively (Phenix was used to superpose the electron density maps, ref 5 in the Supporting Information). In the low-temperature structure of the apoenzyme, access to the binding site of the malonate extender unit is blocked off. In contrast, the room-temperature structure shows a higher-energy conformation that appears to be critical for substrate access. The residue equivalent to Gln9 in the AT from the bacillaene PKS (unpublished structure, Protein Data Bank entry 5dz6) shows hydrogen bonding to a glycerol molecule in the malonate binding site. (b) The catalytic dyad and the residues involved in the substrate specificity are shown as sticks. The residues involved in substrate specificity (Phe190 and Gln9) are colored orange and black for the malonate and citrate complexes, respectively. This picture shows the movement of the Phe190 side chain to accommodate larger substrates in the binding pocket.
