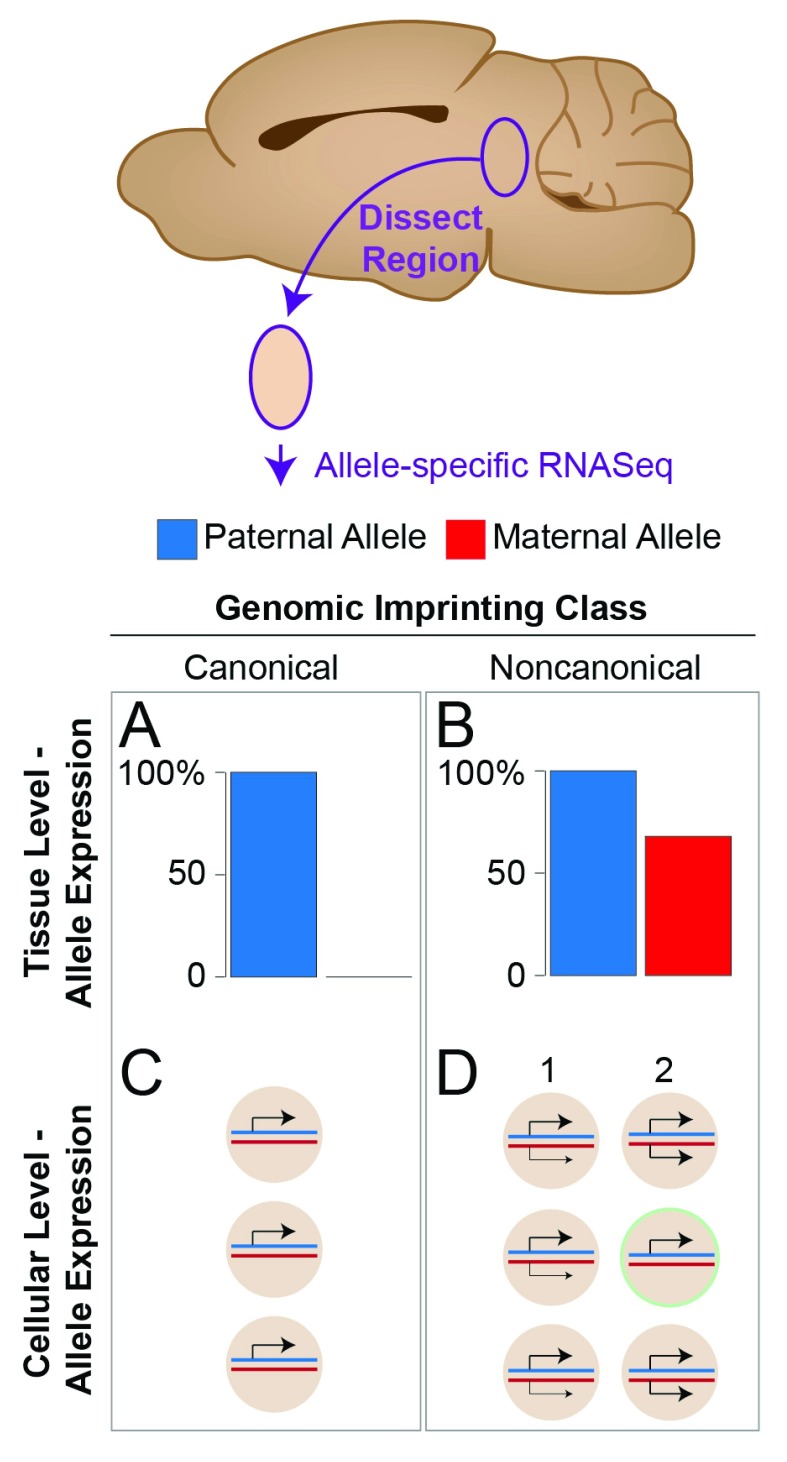
Figure 1. Schematic depiction of canonical versus noncanonical genomic imprinting identified in the mouse.
Canonical and noncanonical imprinting was characterized in different mouse tissues by using RNA-Seq in which allele expression was profiled in a piece of tissue dissected from the brain or in another tissue. ( A) In this chart, canonical imprinting manifests as complete silencing of one parent’s allele (silent maternal allele shown). ( B) In contrast, noncanonical imprinting manifests as a significant bias to express one parental allele at a higher level than the other parental allele (paternal allele bias shown). ( C) At the cellular level, canonical imprinting involves complete silencing of one allele in all cells expressing the gene. ( D) Noncanonical imprinting may involve either (1) an allelic bias in each cell or (2) allele silencing in a subpopulation of cells in the tissue. Distinguishing between these models (1 versus 2) is an active area of research.