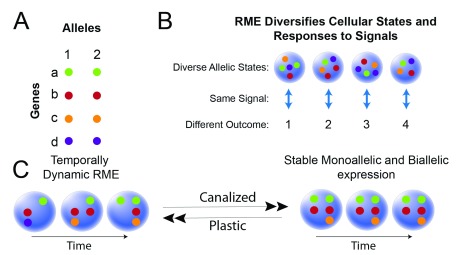
Figure 4. Possible functions for different forms of random monoallelic expression (RME) in promoting cellular diversity and plasticity in mammalian cells.
( A) Schematic of two alleles (1 and 2) for four genes (a–d). ( B) A population of four cells is diversified by clonal or dynamic RME such that each cell expresses a different allelic combination. Exposure of the population to a signal in the environment is predicted to result in different outcomes for each cell (1, 2, 3, 4). Some cells will be in a state that responds better (more quickly or correctly) to the signal than other cells. ( C) Cellular and gene transitions from a temporally dynamic RME state to a stable biallelic or monoallelic (clonal RME or imprinting) state are predicted to shape plasticity versus canalization for gene networks within a cell. The temporally dynamic RME state for different genes is a predicted state of increased plasticity because the cell has access to different allelic combinations that are maintained in a poised state, but these combinations are no longer available once a gene and cell commit to a stable allelic expression state.