Abstract
Noise induced hearing loss (NIHL), a multifactorial disease involving both genetic and environmental factors, is one of the most important occupational health hazards. Nonetheless, the influence of FOXO3 variants on NIHL risk have not been illuminated. This research was conducted to explore the effects of FOXO3 polymorphisms on individual susceptibility to NIHL. A total of 2689 industrial workers from one textile factory of east China were recruited to participate in the current research. Venous blood was collected, questionnaire and pure-tone audiometry (PTA) was conducted by specialist physicians. Then, we performed genotyping of three selected SNPs (rs2802292, rs10457180, and rs12206094) in FOXO3 gene in 566 NIHL patients and 566 controls. Subsequently, the main effects of genotype and its interactions were evaluated. Our results revealed that individuals with the G allele of rs2802292, G allele of rs10457180, T allele of rs12206094 (OR = 1.43, 1.43, and 1.31 respectively) and the haplotype GAC and others (TGT/GGT/GGC/GAT) (rs2802292-rs10457180-rs12206094) (OR = 1.49 and 2.09 respectively) are associated with an increased risk of NIHL in a Chinese population. Stratified analysis showed that an increased NIHL risk was found in the subjects who exposed to noise >16 years with rs2802292 GG/GT and rs10457180 AG/GG genotype with an OR of 1.62 and 1.66 respectively. Multifactor dimensionality reduction analysis indicated that rs10457180, rs2802292, and rs12206094 have interactions and are related to increased NIHL risk (OR = 1.53). The genetic polymorphism rs2802292, rs10457180, and rs12206094 within FOXO3 gene are associated with an increased risk of NIHL in a Chinese population and have potential to be biomarkers for noise exposed workers.
Introduction
Occupational noise is one of the most common occupational hazards for the health of industrial workers, and noise induced hearing loss (NIHL) is the second most frequent form of sensorineural hearing impairment besides age-related hearing loss (ARHL) worldwide[1]. It has been verified that NIHL is a kind of multifactorial disease resulting from the interactions of both genetic and environmental factors[2]. Currently, the mechanism of NIHL has not been completely understood. The possible etiopathogenesis may involve the inner ear cell apoptosis or necrosis caused by oxidative stress or the metabolic products generated during signal transduction and direct mechanical injury to the structures of the cochlea[3–5]. However, numerous population studies have indicated that the subjects had various degrees of NIHL risk even if they were exposed to equal noise intensity level[1, 6]. Animal experiments also prove that genetic variations contribute to the incidence of NIHL[7, 8]. Previous studies have found that single nucleotide polymorphism (SNPs) in HSP70, EYA4, CDH23, GRHL2 and DFNA5 genes are associated with human genetic susceptibility to NIHL and could increase or decrease the risk of NIHL by interaction with occupational noise[9–11]. All the evidence implicates that the genetic susceptibility and its interaction with environmental factors might play an important role in the occurrence and development of NIHL.
Forkhead Box O3 (FOXO3) is a winged helix transcription factor that has been known to regulate longevity in various species including humans and mice[12–14]. FOXO3 regulates the expression of stress response proteins[15] and FOXO3 effectors may ameliorate oxidative stress, block mitosis, induce apoptosis, or promote inflammation[16]. It has been shown that mice are susceptible to adult-onset hearing loss with the hallmark characteristics of auditory neuropathy, namely, elevated auditory thresholds combined with normal outer hair cell function if lacking the transcription factor Foxo3[17]. Also, Foxo3 was found to be necessary for auditory function after noise exposure in a mouse model system and outer hair cells are lost throughout the middle and higher frequencies in absence of Foxo3[18].
However, associations between NIHL and FOXO3 SNPs and their functional significance in the FOXO3 gene were not reported before. In consideration of the vital functions of FOXO3 in hearing maintenance, we speculated that the polymorphisms in FOXO3 gene might have associations with the genetic susceptibility to NIHL. Herein, a case-control study was performed to elucidate the associations between three FOXO3 SNPs, namely rs2802292, rs10457180, and rs12206094, with the genetic susceptibility of NIHL.
Materials and methods
Subjects
The subjects of the current study were all industrial employees from one textile factory of east China who received annual health examinations which were performed by the Jiangsu Provincial Center for Disease Prevention and Control in 2015. A total amount of 2689 people participated in health examinations. Before the investigation, informed consent was obtained from each participant. This study was approved by the Institutional Review Board of Jiangsu Provincial Center for Disease Prevention and Control. Occupational health examination items mainly included venous blood collection, general physical examination and pure-tone audiometry (PTA). During the health examination, personal medical history, smoking, drinking habits and habitual use of drugs were queried. Subjects were excluded from this study according to the following criterions: a) workers with diseases that may affect hearing thresholds (e.g. otitis media, cholesteatoma, ear canal stenosis, etc.); b) workers who have used or are using ototoxic drugs (e.g. aspirin, quinolones, aminoglycosides, etc.). Eventually, 2477 subjects conformed to our criterions.
PTA and NIHL assessment
Audiometry was performed for each participant only after stopping noise exposure for up to12 hours. By using a Madsen Voyager 522 audiometer (Madsen, Taastrup, Denmark), pure tone audiometry was conducted by qualified doctors in a soundproof room.
Definitions of NIHL and control subjects
Hearing loss and normal hearing are identified according to China diagnostic criteria of occupational noise induced deafness (GBZ 49–2007). In this study, occupational noise exposure is identified as levels of noise exposure (Lex) are at least 85 dB (A) for a nominal 8-hour working day. Hearing loss was identified as the binaural hearing thresholds which exceed 25 dB at both high (3000, 4000, 6000 Hz) and speech (500, 1000, 2000 Hz) frequencies. Correspondingly, normal hearing means that the binaural hearing thresholds are under 25 dB both at high and speech frequencies. Hearing thresholds were obtained from the results of PTD. The subjects were divided into two groups: NIHL patient group (noise exposed individuals with hearing loss) and control group (noise exposed individuals with normal hearing). We first selected the NIHL patients, and controls were frequency-matched to the patients by sex, age, and intensity of noise exposure [19]. Eventually, 566 NIHL patients and 566 controls were selected from all eligible subjects.
DNA extractions
Peripheral blood (3 mL) was collected in ethylene diamine tetra acetic acid (EDTA) and taken for DNA isolation and genotyping. DNA was extracted from blood samples of subjects by using the QIAcube HT and QIAamp 96 DNA QIAcube HT Kit (Qiagen, Dusseldorf, Germany) following the manufacturer’s protocol and then stored at -20°C until use.
SNP selection and genotyping
Target SNPs in the FOXO3 genes were selected on the basis of the 1000 Genomes Project database and previous findings from the literature. The criteria for searching for SNPs were as follows: a) MAF (minor allele frequency) of CHB > 0.10; b) a linkage disequilibrium value of r2 >0.8. SNPs of which DNA sequence are not suitable for PCR primer design were excluded. In the end, eleven candidate SNPs were selected using these criteria by Haploview software. Then, we searched these candidate SNPs in Pubmed and found that rs2802292, rs10457180 and rs12206094 were the most commonly reported SNPs of FOXO3 in previous findings. Ultimately three SNPs in the FOXO3 gene were selected for genotyping: rs2802292, rs10457180, and rs12206094.
Genotypes for the selected polymorphisms were screened with the ABI TaqMan SNP genotyping assays (Applied Biosystems, Foster City, Calif., USA) by using the pre-designed commercial genotyping assays. The extracted DNA and genotyping assays were added to TaqMan universal PCR master mix (Roche, Branchburg, N.J., USA) according to the manufacturer’s instructions. The genotyping procedures were then performed by ABI 7900 real-time PCR system (Applied Biosystems). The results were analyzed using ABI 7900 System sequence detection software version 1.2.3 (Applied Biosystems).
Statistical analyses
Statistical analyses was performed by using SPSS 23.0 software (Chicago, IL, USA). The goodness-of-fit χ2 tests were conducted for the Hardy-Weinberg equilibrium rule of the SNPs in FOXO3 gene among the control subjects. Categorical variables are represented as percentages and continuous variables are described as the mean ± SD. Odds ratios (ORs) and 95% confidence interval (95%CI) for genotypes were achieved under conditional logistic regression models adjusted for age, sex, smoking, and drinking. Differences in allele-specific promoter activity and gene expression were compared by Student’s t-test or paired t-test. Haplotype analysis of the polymorphisms was performed using the SHEsis platform [20]. All the P values were corrected (Pc) with Bonferroni corrections and P < 0.05 was used as the criterion of statistical significance.
Results
Demographic characteristics of study subjects and Hardy-Weinberg test of selected SNPs
The general characteristics (age, sex, smoking, drinking, work time with noise and exposure level with noise) and high-frequency hearing threshold of the NIHL cases and controls were shown in Table 1. No significant differences in general characteristics were shown between NIHL cases and controls (P > 0.05). Results also show that there was statistically significant difference between NIHL cases and controls in terms of high-frequency hearing threshold. The average high-frequency hearing threshold was higher for patients (35.77± 9.86) than for controls (14.02± 4.17) (P < 0.001). General information of selected SNPs and results of Hardy-Weinberg test were shown in Table 2. Rs2802292, rs10457180, and rs12206094 are located in 5’ UTR variant, intron variant, intron variant (upstream variant 2KB) of FOXO3 gene respectively and χ2 tests revealed that all SNPs were in the balance of Hardy-Weinberg (P>0.05).
Table 1. Demographic characteristics of study subjects.
| Variables | Cases (n = 566) | Controls (n = 566) | P | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Age (years) | 0.879a | ||||
| Mean±SD | 40.52±6.26 | 40.31±5.85 | 0.556b | ||
| ≤ 35 | 133 | 23.5 | 138 | 24.4 | |
| 35–45 | 320 | 56.5 | 321 | 56.7 | |
| > 45 | 113 | 20.0 | 107 | 18.9 | |
| Sex | 0.646a | ||||
| Male | 527 | 93.1 | 523 | 92.4 | |
| Female | 39 | 6.9 | 43 | 7.6 | |
| Smoking | 0.263a | ||||
| Now | 331 | 58.5 | 315 | 55.7 | |
| Ever | 11 | 1.9 | 19 | 3.4 | |
| Never | 224 | 39.6 | 232 | 41.0 | |
| Drinking | 0.898a | ||||
| Now | 233 | 41.2 | 235 | 41.5 | |
| Ever | 10 | 1.8 | 12 | 2.1 | |
| Never | 323 | 57.1 | 319 | 56.4 | |
| Work time with noise (years) | 0.237a | ||||
| Mean±SD | 18.69±7.59 | 18.06±7.03 | 0.145b | ||
| ≤ 16 | 248 | 43.8 | 261 | 46.1 | |
| > 16 | 318 | 56.2 | 305 | 53.9 | |
| Expose level with noise (dB) | 0.712a | ||||
| Mean±SD | 87.18±7.72 | 87.39±7.39 | 0.651b | ||
| ≤ 85 | 253 | 44.7 | 249 | 44.0 | |
| 85–92 | 109 | 19.3 | 101 | 17.8 | |
| > 92 | 204 | 36.0 | 216 | 38.2 | |
| High frequency hearing threshold shift (dB) | <0.001a | ||||
| Mean±SD | 35.77±9.86 | 14.02±4.17 | <0.001b | ||
| ≤ 26 | 59 | 10.4 | 566 | 100.0 | |
| > 26 | 507 | 89.6 | 0 | 0.0 | |
a Two-sided χ2 test
b Students’ t-test.
Table 2. General information of selected SNPs and Hardy-Weinberg test.
| SNP | Alleles | Chromosome | Functional Consequence | MAF | P for HWE b | |
|---|---|---|---|---|---|---|
| Control | Database a | |||||
| rs2802292 | G/T | 6:108587315 | 5’ UTR variant | 0.212 | 0.189 | 0.172 |
| rs10457180 | A/G | 6:108643836 | Intron variant | 0.205 | 0.200 | 0.754 |
| rs12206094 | C/T | 6:108584997 | Intron variant (upstream variant 2KB) | 0.143 | 0.146 | 0.858 |
a Data from NCBI dbSNP
b P value of Hardy-Weinberg test.
Multivariate analyses of FOXO3 SNPs with the risk of NIHL
Three FOXO3 SNPs (rs2802292, rs10457180, and rs12206094) were selected to genotype in 1132 noise exposed workers (566 NIHL patients and 566 controls). Table 3 presents the results of genotype and allele distributions of rs2802292, rs10457180, and rs12206094 in codominant, dominant, recessive and allelic model. Results are shown that the genotype frequencies of rs2802292, rs10457180, and rs12206094 in codominant model among cases and controls were statistically significantly different (P = 0.001, 0.001 and 0.046 respectively). In the dominant model, the rs2802292 GT+GG and rs10457180 AG+GG were significantly associated with NIHL risk (P = 0.008 and 0.005 respectively). Subsequent logistic regression analysis adjusting for age, sex, smoking and drinking showed that individuals with rs2802292 GT+GG and rs10457180 AG+GG had increased NIHL risk with OR of 1.40 (95%CI = 1.10–1.78) and 1.44 (95%CI = 1.13–1.82). In the recessive model, the rs2802292 GG (OR = 2.24, 95% CI = 1.38–3.36), rs10457180 GG (OR = 2.16, 95% CI = 1.29–3.61) and rs12206094 TT (OR = 2.10, 95% CI = 1.04–4.24) genotypes conferred a significantly increased risk for NIHL (P = 0.001, 0.004, and 0.043 respectively). In the allelic model, the rs2802292 G (OR = 1.43, 95% CI = 1.18–1.74), rs10457180 G (OR = 1.43, 95% CI = 1.18–1.75) and rs12206094 T (OR = 1.31, 95% CI = 1.04–1.64) alleles conferred a significantly increased risk for NIHL (P <0.001, P <0.001, and P = 0.026 respectively). Thus, our data revealed that FOXO3 SNP rs2802292, rs10457180, and rs12206094 may have a significant association with NIHL risk, and those individuals who carry rs2802292 G, rs10457180 G, and rs12206094 T allele may have significantly increased NIIHL susceptibility.
Table 3. Distribution of three polymorphisms and the association with NIHL.
| Genetic models | Genotypes | Cases | Controls | Pa | Adjusted OR | ||
|---|---|---|---|---|---|---|---|
| n = 566 | % | n = 566 | % | (95% CI)b | |||
| rs2802292 | |||||||
| Codominant | TT | 307 | 54.2 | 352 | 62.2 | 0.001 | 1.00 (Ref.) |
| GT | 204 | 36.0 | 188 | 33.2 | 1.26 (0.98–1.02) | ||
| GG | 55 | 9.7 | 26 | 4.6 | 2.44 (1.49–4.00) | ||
| Dominant | TT | 307 | 54.2 | 352 | 62.2 | 0.008 | 1.00 (Ref.) |
| GT/GG | 259 | 45.8 | 214 | 37.8 | 1.40 (1.10–1.78) | ||
| Recessive | TT/GT | 511 | 90.3 | 540 | 95.4 | 0.001 | 1.00 (Ref.) |
| GG | 55 | 9.7 | 26 | 4.6 | 2.24 (1.38–3.63) | ||
| Alleles | T | 818 | 72.3 | 892 | 78.8 | <0.001 | 1.00 (Ref.) |
| G | 314 | 27.7 | 240 | 21.2 | 1.43 (1.18–1.74) | ||
| rs10457180 | |||||||
| Codominant | AA | 309 | 54.6 | 357 | 63.1 | 0.001 | 1.00 (Ref.) |
| AG | 210 | 37.1 | 186 | 32.9 | 1.32 (1.03–1.69) | ||
| GG | 47 | 8.3 | 23 | 4.1 | 2.40 (1.42–4.05) | ||
| Dominant | AA | 309 | 54.6 | 357 | 63.1 | 0.005 | 1.00 (Ref.) |
| AG/GG | 257 | 45.4 | 209 | 36.9 | 1.44 (1.13–1.82) | ||
| Recessive | AA/AG | 519 | 91.7 | 543 | 95.9 | 0.004 | 1.00 (Ref.) |
| GG | 47 | 8.3 | 23 | 4.1 | 2.16 (1.29–3.61) | ||
| Alleles | A | 828 | 73.1 | 900 | 79.5 | <0.001 | 1.00 (Ref.) |
| G | 304 | 26.9 | 232 | 20.5 | 1.43 (1.18–1.75) | ||
| rs12206094 | |||||||
| Codominant | CC | 389 | 68.7 | 416 | 73.5 | 0.046 | 1.00 (Ref.) |
| CT | 152 | 26.9 | 138 | 24.4 | 1.19 (0.91–1.56) | ||
| TT | 25 | 4.4 | 12 | 2.1 | 2.20 (1.09–4.45) | ||
| Dominant | CC | 389 | 68.7 | 416 | 73.5 | 0.088 | 1.00 (Ref.) |
| CT/TT | 177 | 31.3 | 150 | 26.5 | 1.28 (0.99–1.65) | ||
| Recessive | CC/CT | 541 | 95.6 | 554 | 97.9 | 0.043 | 1.00 (Ref.) |
| TT | 25 | 4.4 | 12 | 2.1 | 2.10 (1.04–4.24) | ||
| Alleles | C | 930 | 82.2 | 970 | 85.7 | 0.026 | 1.00 (Ref.) |
| T | 202 | 17.8 | 162 | 14.3 | 1.31 (1.04–1.64) | ||
a Two-sided χ2 test
b Adjusted for age, sex, smoking, drinking in logistic regression model.
Stratified analyses of rs2802292, rs10457180, and rs12206094 polymorphism and NIHL risk
The impacts of rs2802292, rs10457180, and rs12206094 genotypes in NIHL on a series of risk characteristics were analyzed in a dominant model. The results were shown in Table 4. Significant differences were found in the genotype distributions between cases and controls for the group in noise exposure time (>16 years) in rs2802292 and rs10457180 (P = 0.006 and 0.004). After adjusting for age, sex, smoking, and drinking in logistic regression model, an increased NIHL risk was found in the subjects who exposed to noise >16 years with rs2802292 GG/GT and rs10457180 AG/GG genotype with an OR of 1.62 (95% CI = 1.17–2.26) and 1.66 (95% CI = 1.19–2.31). For persons exposed to noise ≤85dB with rs10457180 AG/GG have an increased risk for NIHL (OR = 1.81, 95% CI = 1.25–2.61, P = 0.003).
Table 4. Stratified analysis of SNPs in a dominant model.
| SNPs | Group | Genotype | Work time with noise (years) | Expose level with noise (dB) | |||
|---|---|---|---|---|---|---|---|
| ≤16 | > 16 | ≤85 | 85–92 | > 92 | |||
| rs2802292 | case | GG/GT | 113 | 146 | 112 | 52 | 95 |
| TT | 135 | 172 | 141 | 57 | 109 | ||
| control | GG/GT | 107 | 107 | 89 | 33 | 92 | |
| TT | 154 | 198 | 160 | 68 | 124 | ||
| Pa | 0.298 | 0.006 | 0.051 | 0.027 | 0.413 | ||
| Adjusted OR (95% CI)b | 1.22 (0.85–1.74) | 1.62 (1.17–2.26) | 1.50 (1.04–2.16) | 1.77 (1.00–3.13) | 1.18 (0.80–1.75) | ||
| rs10457180 | case | AG/GG | 115 | 142 | 117 | 49 | 91 |
| AA | 133 | 176 | 136 | 60 | 113 | ||
| control | AG/GG | 107 | 102 | 83 | 32 | 94 | |
| AA | 154 | 203 | 166 | 69 | 122 | ||
| Pa | 0.222 | 0.004 | 0.003 | 0.048 | 0.822 | ||
| Adjusted OR (95% CI)b | 1.27 (0.89–1.81) | 1.66 (1.19–2.31) | 1.81 (1.25–2.61) | 1.73 (0.97–3.09) | 1.06 (0.72–1.57) | ||
| rs12206094 | case | CT/TT | 80 | 97 | 77 | 33 | 67 |
| CC | 168 | 221 | 176 | 76 | 137 | ||
| control | CT/TT | 78 | 72 | 62 | 18 | 70 | |
| CC | 183 | 233 | 187 | 83 | 146 | ||
| Pa | 0.563 | 0.053 | 0.166 | 0.035 | 0.924 | ||
| Adjusted OR (95% CI)b | 1.14 (0.78–1.66) | 1.47 (1.02–2.10) | 1.39 (0.93–2.07) | 1.87 (0.96–3.65) |
1.02 (0.67–1.54) | ||
a Two-sided χ2 test.
b Adjusted for age, sex, smoking, drinking in logistic regression model.
Association between the haplotypes of FOXO3 SNPs with NIHL risk
Furthermore, analysis of the haplotype frequencies of the three SNPs was performed between NIHL cases and controls (Table 5). Four common haplotypes (frequency > 5%) derived from the three SNPs accounting for 90% of the haplotype variations were selected and the rest of haplotypes were pooled into the mixed group. Ultimately, the haplotype GAC and others (TGT/GGT/GGC/GAT) (rs2802292-rs10457180-rs12206094) in FOXO3 gene were found to be associated with an increased risk for NIHL (OR = 1.49 and 2.09) compared to TAC haplotype.
Table 5. Frequencies of inferred haplotypes among the cases and controls and their association with risk NIHL.
| Haplotypesa | Case (n = 566) | Control (n = 566) | Pb | Adjusted OR | Global Pd | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | (95% CI)c | |||
| TAC | 610 | 53.9 | 697 | 61.6 | 1.00 (Ref.) | <0.001 | |
| GAC | 209 | 18.5 | 190 | 16.8 | 0.039 | 1.27 (1.01–1.59) | |
| TGT | 130 | 11.5 | 118 | 10.4 | 0.080 | 1.28 (0.97–1.68) | |
| TGC | 72 | 6.4 | 66 | 5.8 | 0.209 | 1.25 (0.88–1.78) | |
| Otherse | 111 | 9.8 | 61 | 5.4 | <0.001 | 2.09 (1.50–2.91) | |
a The alleles of haplotypes were arrayed as rs2802292-rs10457180-rs12206094.
b Two-sided χ2 test.
c Adjusted for age, sex, smoking, drinking in logistic regression model.
d Generated by permutation test with 1000 times of simulation.
e Haplotypes (TGT/GGT/GGC/GAT) were pooled into the mixed group.
Comparison of high-frequency hearing threshold shift of three SNPs genotypes
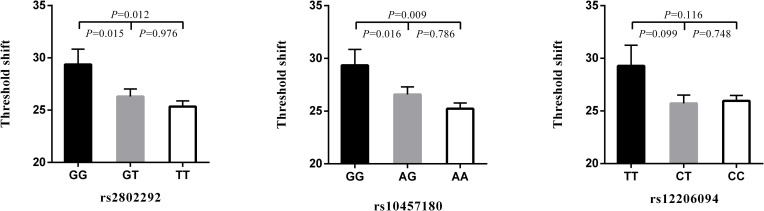
Fig 1 shows the results of the comparison of high-frequency hearing threshold shift of rs2802292, rs10457180, and rs12206094 genotypes in 1132 noise exposed workers. Subjects with GG genotype of rs2802292 have a significantly higher high-frequency hearing threshold shift than people with GT and TT genotype (P = 0.015 and 0.012). Persons with GG genotype of rs10457180 were also found to have higher high-frequency hearing threshold shift than people with AG and AA genotype (P = 0.016 and 0.009).
Fig 1. Comparison of high-frequency hearing threshold shift of three SNPs.
Comparison of high-frequency hearing threshold shift of rs2802292, rs10457180, and rs12206094 genotypes in all subjects. Data are presented as mean ± SE and analyzed by ANOV.
Multifactor dimensionality reduction analyses of the interaction between the three SNPs
MDR analysis results of the interaction between the three SNPs were presented in Table 6. The interaction results suggested that the rs10457180-rs2802292-rs12206094 model got the highest cross-validation consistency and testing balanced accuracy and was related to increased NIHL risk (OR = 1.53, 95%CI = (1.20–1.94), P = 0.0005).
Table 6. MDR analysis results of the interaction between the three SNPs.
| Model | Training balanced accuracy | Testing balanced accuracy | Cross-validation consistency | P | OR(95%CI) |
|---|---|---|---|---|---|
| rs10457180 | 0.5426 | 0.538 | 9/10 | 0.0037 | 1.42(1.12–1.80) |
| rs10457181-rs2802292 | 0.5463 | 0.5274 | 7/10 | 0.0016 | 1.47(1.16–1.87) |
| rs10457181-rs2802292-rs12206094 | 0.5507 | 0.538 | 10/10 | 0.0005 | 1.53(1.20–1.94) |
Discussion
Single nucleotide polymorphism (SNP) is the one of the most common types of genetic variant in the human genome and there are about fifteen million SNPs in all human group[21]. Haplotype, defined as a specific set of alleles observed on a single chromosome, or a part of a chromosome, has been an integral part of human genetics for decades[22]. However, the genomic distribution of SNPs is not homogenous; most of the SNPs occur in noncoding regions of gene more frequently than in coding regions. At present, detection methods of SNP include denaturing gradient gel electrophoresis (DDGE), single-strand conformational polymorphism (SSCP), cleaved amplified polymorphic sequence (CAPS), denaturing gradient gel electrophoresis (DDGE), and allele-specific PCR (such as TaqMan SNP genotype-PCR).
In the current study, a genetic association analysis was performed on three selected FOXO3 SNPs (rs2802292, rs10457180, and rs12206094) in 566 NIHL patients and 566 controls by TaqMan SNP genotyping assay. Results reveal that rs2802292 G allele, rs10457180 G allele, and rs12206094 T allele in FOXO3 are associated with a significantly higher risk of NIHL. Subsequent haplotype analysis shows that the haplotype GAC and others (TGT/GGT/GGC/GAT) (rs2802292-rs10457180-rs12206094) confers increased risks of NIHL. These findings support our hypothesis that the FOXO3 polymorphism may contribute to susceptibility to NIHL. As far as we know, this may be the first association study showing that FOXO3 gene is correlated with an increased risk of NIHL in a Chinese population.
Recent research has shown the essential role of transcription factor FOXO3 in the cochlea in hearing maintenance[17]. Foxo3 protein is detected in the nuclei of spiral ganglion neurons, hair cells, and pillar cells of young adult mice suggesting that it acts in sensory cells. Notably, Foxo3 protein becomes localized to the cytoplasm of spiral ganglion neurons as mice age. Exposure to nondamaging noise levels drives Foxo3 nuclear localization indicating that higher cochlear activity levels promote Foxo3-dependent transcription by increasing Foxo3 concentration near DNA[17]. Moreover, Foxo3 can drive transcription of the master mitochondrial regulator Pgc-1α. Pgc-1α coordinates new production of mitochondrial proteins from both the nucleus and mitochondrial DNA (mtDNA)[23]. In conjunction with Pgc-1α, Foxo3 directly interacts with the promoters of Sod2, catalase, and peroxiredoxin to drive their expression[23]. Sod2, catalase, and peroxiredoxin which contains free radicals have been known to have an essential role in the cochlea in noise-induced hearing loss (NIHL)[5]. Foxo3 is required for auditory function after noise exposure in a mouse model system and absent Foxo3, outer hair cells are lost throughout the middle and higher frequencies[18]. Above evidence support an influential role of transcription factor Foxo3 in contributing to the maintenance and protection of hearing.
Despite all the three SNPs selected locate in noncoding region of FOXO3 gene, numerous studies have shown that functional consequences of noncoding SNPs involve in the regulation of protein-coding genes and lncRNA regulation[24, 25]. Genetic variation at FOXO3 rs12212067 has been proved to drive allele-specific expression of FOXO3 and modulate inflammatory cytokine production in monocytes through a paracrine TGFβ1-dependent mechanism[26]. Also, Foxo3 intron variations may alter the dysregulated biogenesis of circ-Foxo3. Ectopic expression of circ-Foxo3 could bind to the cell cycle proteins cyclin-dependent kinase 2 and cyclin-dependent kinase inhibitor 1, resulting in the formation of a ternary complex which arrests the function of CDK2 and blocked cell cycle progression[27].
Our current study has several potential limitations. (i) The sample size of our study was relatively larger compared to previous research. However, the power of statistical tests may not be fully enough due to the lower biological effects of an individual SNP. Therefore, large sample size and cohort studies are needed in future to confirm the effects of the FOXO3 polymorphism on NIHL; (ii) Study subjects of this case-control study are limited by the Chinese population. Thus, our results may likely be better generalized to Chinese Han population and limit external generalizability; (iii) The selected SNPs are noncoding, thus we assume these variants to be linked with one or more functional variants within the FOXO3 gene or its regulatory regions. Future fine mapping of the FOXO3 gene may detect such functional variants; (iv) The loss of connections between inner hair cells and auditory nerve has been proved to be associated with loss of FOXO3 by animal studies and could change suprathresholds audition. This "hidden hearing loss" is not always detected with the routine tests of changes in hearing thresholds by pure tone audiometry. As this is a cohort study, suprathreshold audiometry is necessary in the follow-up examination of these noise exposed workers to detect "hidden hearing loss" and verify the role of FOXO3 gene variation in noise induced hearing loss.
In conclusion, rs2802292 G allele, rs10457180 G allele, and rs12206094 T allele in FOXO3 are associated with a significantly higher risk of NIHL. These three noncoding SNPs may involve in the regulation of protein-coding genes, lncRNA, and circRNA mechanism of FOXO3. Thus, these findings show that FOXO3 SNPs (rs2802292, rs10457180, and rs12206094) may have important roles in noise induced hearing loss and have potential to be biomarkers for noise exposed workers.
Supporting information
Demographic characteristics and genotype result of each subject.
(XLS)
Acknowledgments
We thank all noise-exposed workers for participating in the study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Baoli Zhu was funded by Jiangsu Province’s Outstanding Medical Academic Leader program (CXTDA2017029) and Huanxi Shen was funded by Jiangsu Program for Young Medical Talents (QNRC2016528).
References
- 1.Konings A, Van Laer L, Van Camp G. Genetic studies on noise-induced hearing loss: a review. Ear Hear. 2009;30(2):151–9. doi: 10.1097/AUD.0b013e3181987080 . [DOI] [PubMed] [Google Scholar]
- 2.Carlsson PI, Van Laer L, Borg E, Bondeson ML, Thys M, Fransen E, et al. The influence of genetic variation in oxidative stress genes on human noise susceptibility. Hear Res. 2005;202(1–2):87–96. doi: 10.1016/j.heares.2004.09.005 . [DOI] [PubMed] [Google Scholar]
- 3.Henderson D, Bielefeld EC, Harris KC, Hu BH. The role of oxidative stress in noise-induced hearing loss. Ear Hear. 2006;27(1):1–19. doi: 10.1097/01.aud.0000191942.36672.f3 . [DOI] [PubMed] [Google Scholar]
- 4.Le Prell CG, Dolan DF, Schacht J, Miller JM, Lomax MI, Altschuler RA. Pathways for protection from noise induced hearing loss. Noise & health. 2003;5(20):1–17. NO_DOI. . [PubMed] [Google Scholar]
- 5.Le Prell CG, Yamashita D, Minami SB, Yamasoba T, Miller JM. Mechanisms of noise-induced hearing loss indicate multiple methods of prevention. Hear Res. 2007;226(1–2):22–43. doi: 10.1016/j.heares.2006.10.006 ; PubMed Central PMCID: PMC1995566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson D, Subramaniam M, Boettcher FA. Individual susceptibility to noise-induced hearing loss: an old topic revisited. Ear Hear. 1993;14(3):152–68. . [DOI] [PubMed] [Google Scholar]
- 7.White CH, Ohmen JD, Sheth S, Zebboudj AF, McHugh RK, Hoffman LF, et al. Genome-wide screening for genetic loci associated with noise-induced hearing loss. Mammalian genome: official journal of the International Mammalian Genome Society. 2009;20(4):207–13. doi: 10.1007/s00335-009-9178-5 ; PubMed Central PMCID: PMC2823581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis RR, Newlander JK, Ling X, Cortopassi GA, Krieg EF, Erway LC. Genetic basis for susceptibility to noise-induced hearing loss in mice. Hear Res. 2001;155(1–2):82–90. . [DOI] [PubMed] [Google Scholar]
- 9.Kowalski TJ, Pawelczyk M, Rajkowska E, Dudarewicz A, Sliwinska-Kowalska M. Genetic variants of CDH23 associated with noise-induced hearing loss. Otology & neurotology: official publication of the American Otological Society, American Neurotology Society [and] European Academy of Otology and Neurotology. 2014;35(2):358–65. doi: 10.1097/MAO.0b013e3182a00332 . [DOI] [PubMed] [Google Scholar]
- 10.Chang NC, Ho CK, Lin HY, Yu ML, Chien CY, Ho KY. Association of polymorphisms of heat shock protein 70 with susceptibility to noise-induced hearing loss in the Taiwanese population. Audiology & neuro-otology. 2011;16(3):168–74. doi: 10.1159/000317119 . [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Liu Y, Zhang L, Yang Z, Yang L, Wang X, et al. Associations of genetic variations in EYA4, GRHL2 and DFNA5 with noise-induced hearing loss in Chinese population: a case- control study. Environmental health: a global access science source. 2015;14:77 doi: 10.1186/s12940-015-0063-2 ; PubMed Central PMCID: PMC4581404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429(6991):562–6. doi: 10.1038/nature02549 . [DOI] [PubMed] [Google Scholar]
- 13.Shimokawa I, Komatsu T, Hayashi N, Kim SE, Kawata T, Park S, et al. The life-extending effect of dietary restriction requires Foxo3 in mice. Aging cell. 2015;14(4):707–9. doi: 10.1111/acel.12340 ; PubMed Central PMCID: PMC4531086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, et al. FOXO3A genotype is strongly associated with human longevity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(37):13987–92. doi: 10.1073/pnas.0801030105 ; PubMed Central PMCID: PMC2544566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer EL, Brunet A. FOXO transcription factors in ageing and cancer. Acta physiologica. 2008;192(1):19–28. doi: 10.1111/j.1748-1716.2007.01780.x . [DOI] [PubMed] [Google Scholar]
- 16.Hwang JW, Rajendrasozhan S, Yao H, Chung S, Sundar IK, Huyck HL, et al. FOXO3 deficiency leads to increased susceptibility to cigarette smoke-induced inflammation, airspace enlargement, and chronic obstructive pulmonary disease. Journal of immunology. 2011;187(2):987–98. doi: 10.4049/jimmunol.1001861 ; PubMed Central PMCID: PMC3131437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilels F, Paquette ST, Zhang J, Rahman I, White PM. Mutation of Foxo3 causes adult onset auditory neuropathy and alters cochlear synapse architecture in mice. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33(47):18409–24. doi: 10.1523/JNEUROSCI.2529-13.2013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilels F, Paquette ST, Beaulac HJ, Bullen A, White PM. Severe hearing loss and outer hair cell death in homozygous Foxo3 knockout mice after moderate noise exposure. Scientific reports. 2017;7(1):1054 doi: 10.1038/s41598-017-01142-3 ; PubMed Central PMCID: PMC5430619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen H, Cao J, Hong Z, Liu K, Shi J, Ding L, et al. A functional Ser326Cys polymorphism in hOGG1 is associated with noise-induced hearing loss in a Chinese population. PLoS One. 2014;9(3):e89662 doi: 10.1371/journal.pone.0089662 ; PubMed Central PMCID: PMC3943766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–8. doi: 10.1038/sj.cr.7290272 . [DOI] [PubMed] [Google Scholar]
- 21.Jiang R, Duan J, Windemuth A, Stephens JC, Judson R, Xu C. Genome-wide evaluation of the public SNP databases. Pharmacogenomics. 2003;4(6):779–89. doi: 10.1517/phgs.4.6.779.22821 . [DOI] [PubMed] [Google Scholar]
- 22.International HapMap C. The International HapMap Project. Nature. 2003;426(6968):789–96. doi: 10.1038/nature02168 . [DOI] [PubMed] [Google Scholar]
- 23.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–24. doi: 10.1016/S0092-8674(00)80611-X . [DOI] [PubMed] [Google Scholar]
- 24.Meyer KB, Maia AT, O'Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P, et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS genetics. 2011;7(7):e1002165 doi: 10.1371/journal.pgen.1002165 ; PubMed Central PMCID: PMC3140991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo H, Ahmed M, Zhang F, Yao CQ, Li S, Liang Y, et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nature genetics. 2016;48(10):1142–50. doi: 10.1038/ng.3637 . [DOI] [PubMed] [Google Scholar]
- 26.Lee JC, Espeli M, Anderson CA, Linterman MA, Pocock JM, Williams NJ, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155(1):57–69. doi: 10.1016/j.cell.2013.08.034 ; PubMed Central PMCID: PMC3790457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic acids research. 2016;44(6):2846–58. doi: 10.1093/nar/gkw027 ; PubMed Central PMCID: PMC4824104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographic characteristics and genotype result of each subject.
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.