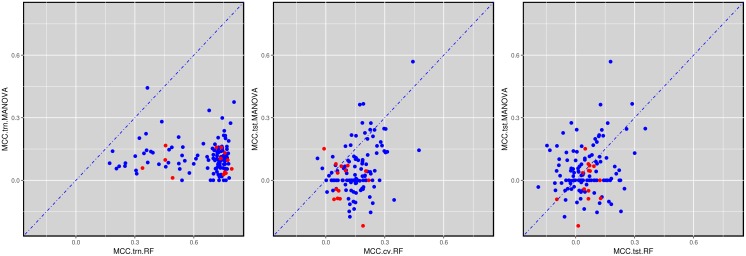
Figure 7. Global performance assessment of single-gene markers versus multi-gene markers across the 127 drugs.
(Left) Performance assessment based on the training data would be strongly biased towards multi-gene markers due to overfitting (99.2% of drugs are better predicted by multi-gene markers). (Middle) The performance of single-gene markers on the test set is compared to the 10-fold cross-validated performance of multi-gene markers using training data. The same cross-validation is also used for model selection (here, identifying the optimal RF’s mtry value) and hence this performance assessment is expected to be biased towards multi-gene markers (85.0% of drugs are better predicted by multi-gene markers). (Right) All the comparable data released after the initial GDSC release was used as a time-stamped test set, which is the most realistic form of retrospective performance assessment. 55.1% of the drugs are now better predicted by their multi-gene marker. Furthermore, 109 drugs are better predicted than random (MCC>0) with multi-gene markers, 66 of these by their multi-gene marker and the remaining 43 by their best single-gene marker. The large difference between these results and that of cross-validated results (55.1% vs 85%) shows that using the same cross-validation for both model selection and its assessment results in an overoptimistic view of predictive performance.