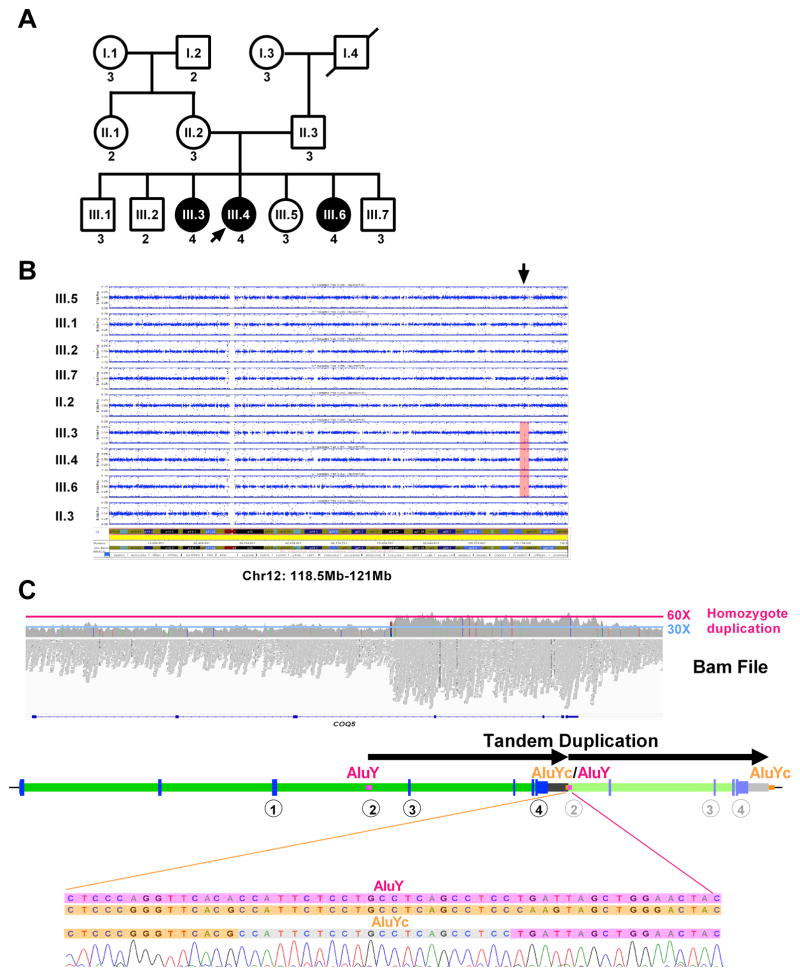
Fig. 1. Patients included in the study.
(A) Family pedigree. Patients who are affected are shown in filled circles. Male individuals are represented by squares, while female individuals are represented by circles. (B) SNP data analysis of all members of the family shows a single homozygous region, from 118.5Mb-121Mb on chromosome 12 (hg19) (arrow).
(C) Whole genome sequencing on III.4 was inconclusive, but analysis of Bam file shows an increase of the number of reads in the candidate locus on chromosome 12 between positions ~120,940,150 and ~120,949,950, with an average number of reads of ~60X compared to the rest of the region (~30X). A cartoon below the Bam file shows the duplication in tandem, breakpoints confirmed by Sanger sequencing), between an AluYc (chr12:120939934–120940228) and an AluY (chr12:120949733–120950042). The encircled numbers below the gene show the positions of the different Taqman probes used for testing the copy number of the duplicated region, which is shown in (A) as the number below each box or circle.