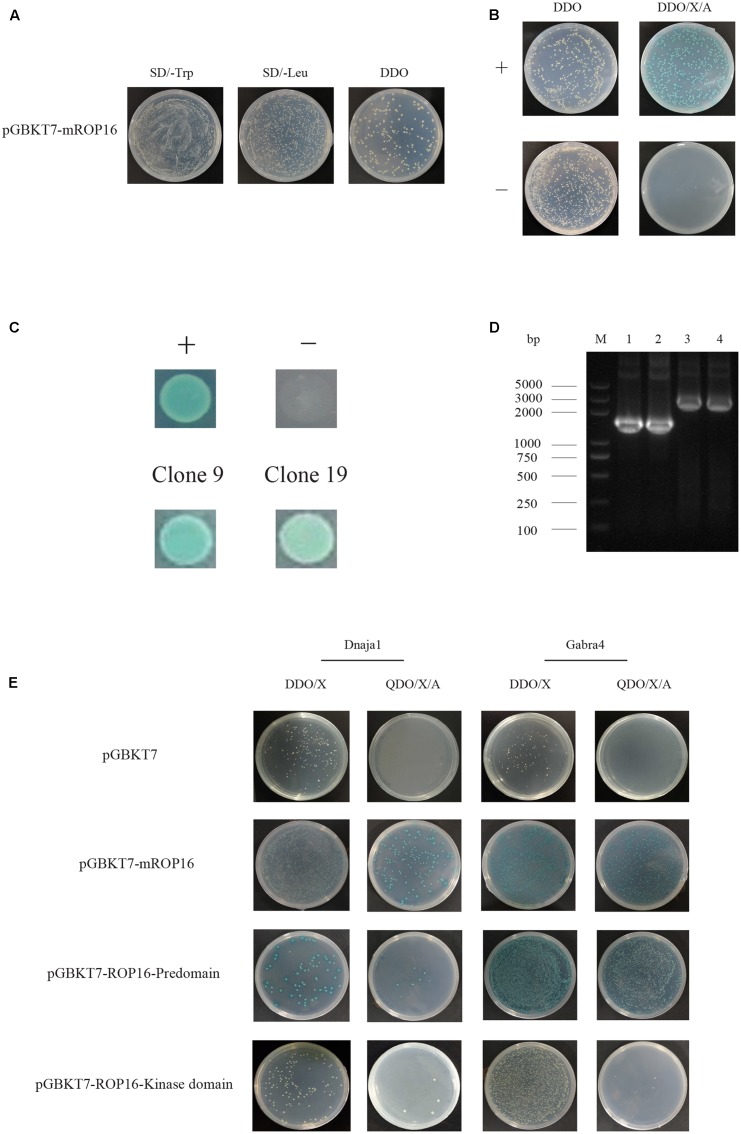
FIGURE 2.
Screen of mouse proteins interacting with mROP16 using yeast two-hybrid (Y2H). (A) Determination of mating efficiency. Equal amount of mated cultures (Y2H Gold/mROP16 + Y187/mouse cDNA library) were plated on SD/-Trp, SD/-Leu, and SD/-Trp/-Leu (DDO) plates. Mating efficiency was calculated by dividing the number of colonies on DDO plate by the number of colonies on SD/-Trp or SD/-Leu plates, whichever is less (In this example, there are less colonies on SD/-Leu plate than on SD/-Trp plate, therefore the number of colonies on SD/-Leu plate was used for mating efficiency calculation). (B) Growth of positive (+, pGBKT7-53 and pGADT7-T) and negative (-, pGBKT7-Lam and pGADT7-T) controls on Y2H DDO and DDO/X/A indication plates. (C) Growth of two positive clones (#9 and #19) on QDO/X/A plates, + and – are positive and negative control colonies, respectively. (D) PCR amplification of the inserts in positive clones. M: DNA Marker. Lane 1–2: Clone 9; Lane 3–4: Clone 19. (E) Interaction between the two hits and different domains of ROP16. Dnaja1 or Gabra4 expressing plasmids and different domains of ROP16 were transformed into Y187 and Y2H Gold cells, respectively. Transformants were used to mate and grown on DDO/X and QDO/X/A plates to monitor protein interactions.