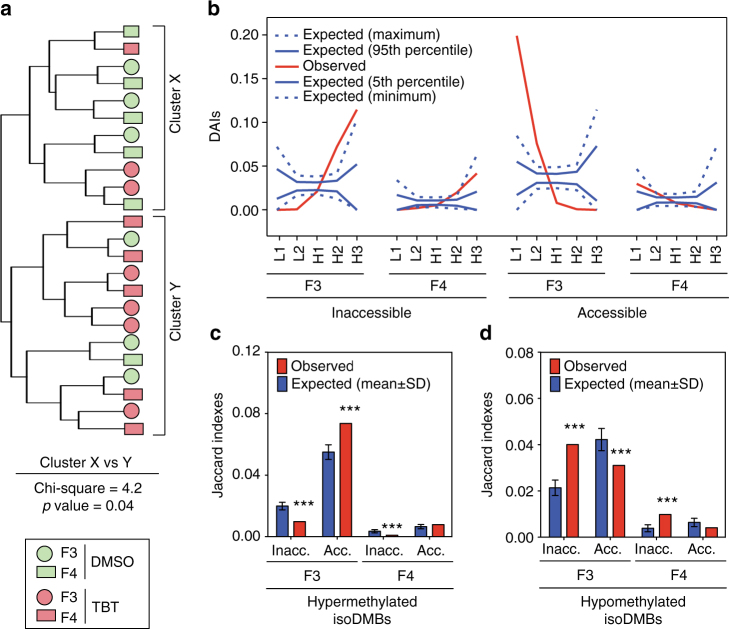
Fig. 5.
Ancestral TBT exposure leads to altered sperm chromatin accessibility. ATAC-seq was used to compare chromatin accessibility patterns (n = 6 per treatment group and generation). a Hierarchical clustering of DMSO and TBT sperm samples using their chromatin accessibility similarities obtained using chromVar guided by H2 isochores. Chi-square tests were used to compare the assortment of DMSO and TBT, or F3 and F4 samples with regard to the two main clusters (X and Y), and subclusters within each cluster (see Methods and Supplementary Data 13). b Distribution of differentially accessible isochores (DAIs) defined using MEDIPS (see Methods for further details) in F3 and F4 generations. DAIs for which ATAC-seq read coverage was larger in TBT than in DMSO samples were deemed as accessible DAIs (Acc.), whereas DAIs for which ATAC-seq read coverage was larger in DMSO than in TBT samples were deemed as inaccessible DAIs (Inacc.). Observed DAI fractions outlying the area defined by 5th and 95th percentiles lines are considered significant (p < 0.05), whereas observed DAI fractions outlying the area defined by minimum and maximum lines are considered highly significant (p < 0.0001). c, d Overlap between DAIs in F3 sperm and hyper and hypomethylated isoDMBs in F4 adipose tissue. The Jaccard index that measures the similarity between datasets was used to quantify the general overlap between F3 and F4 DAIs and F4 adipose isoDMBs (see Methods). Expected Jaccard indexes were calculated after randomly rearranging isoDMBs location 10,000 times respecting isoDMBs, and DAIs length and chromosome assortment using HyperBrowser. p values represent the number of random simulations showing Jaccard indexes more extreme than observed ones. Data is presented as mean ± s.d. **p < 0.01, ***p < 0.001