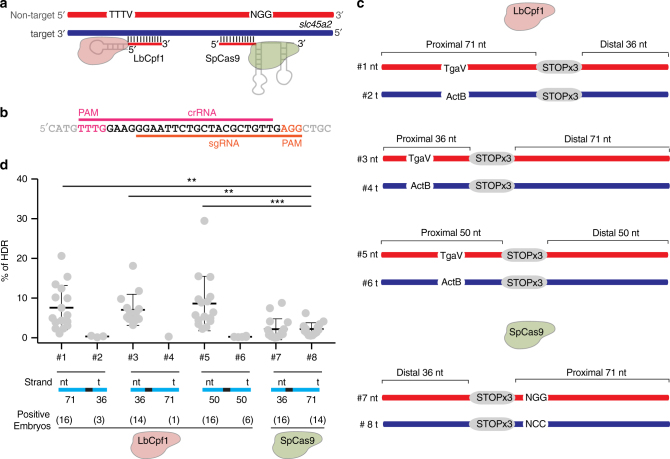
Fig. 4.
LbCpf1-mediated homology-directed repair. a Schematic illustrating LbCpf1–crRNA RNP complex and SpCas9–sgRNA RNP complex interaction with their respective DNA targets. crRNA and sgRNA-DNA annealing occurs on the target strand (blue), and PAM sequences are in the non-target strand (red). b crRNA (pink line) and sgRNA (orange line) overlapping target sequences in the albino locus used for this analysis. c Schema illustrating different donor ssDNA (#1–6) complementary to either the target strand (t) or non-target strand (nt) and with symmetric or asymmetric homology arms used in combination with LbCpf1–crRNA. PAM sequence was modified (TgaV/ActB) to prevent new editing post-HDR. An optimized ssDNA donor (#8) described for SpCas9-induced HDR19 and its complementary version (#7) were used in combination with SpCas9–sgRNA RNP complexes as references for comparison (bottom). d qPCR quantification showing % of HDR from individual embryos when using LbCpf1 and different ssDNA donors in comparison to SpCas9. % of HDR: amount of integrated DNA per total amount of genomic DNA per embryo (see Methods for details). Results are shown as the averages ± S.D. of the means from 16 embryos in two independent experiments (n = 8 embryos per experiment). Positive embryos: number of embryos per condition showing a detectable qPCR amplification signal. The data were analyzed by Kruskal–Wallis test followed by Dunn’s post test for significance vs. control condition (#8), **p < 0.01, ***P < 0.001