Figure 1.
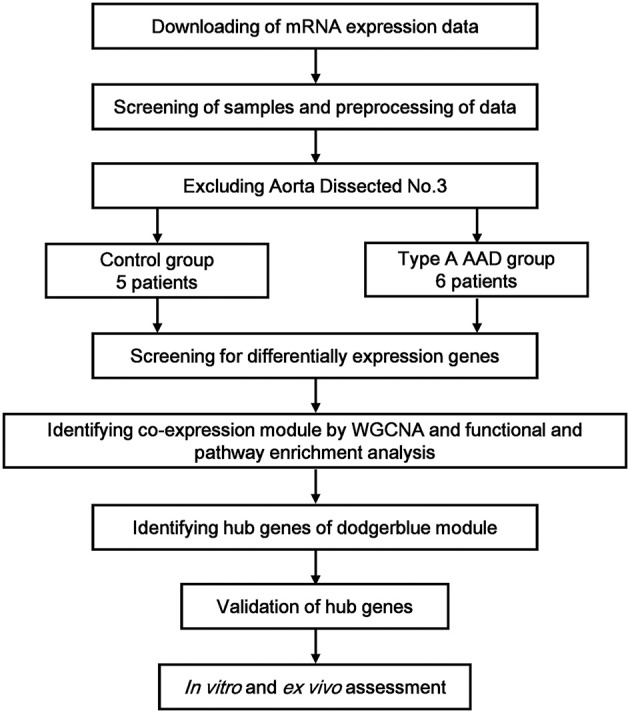
Flow diagram of the study approach. mRNA microarray analyses were performed on aortic specimens obtained from seven AAD patients and five control group (GSE 52093). After excluding Aorta Dissected No. 3 through clustering analysis, six human type A AAD samples and five control samples were included in the subsequent analysis. From among differentially-expressed genes, network analysis of gene expression in aortic dissection identifies 9 modules of co-expression genes. Afterwards, the relevance between each module and aortic dissection were tested through calculating the relevance between the feature vectors of modules and phenotypes, and the dodgerblue module was identified as the most relevant module. Functional and pathway enrichment analysis were performed for the genes in the dodgerblue module. The top 10 genes with the highest connectivity in dodgerblue module were taken as hub genes. Validation quantitative polymerase chain reaction (qRT-PCR) was performed for the 10 hub genes in samples obtained from our institute, including 6 organ donors and 6 AAD patients. At last, EA.hy926 cells and THP-1 cells were used for functional verification in vitro.
