Fig. 1.
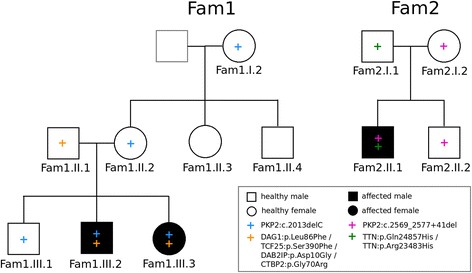
Pedigrees of the two families analyzed in this study. Only labeled individuals were sequenced. +: Heterozygous variant, filled black symbols: affected individuals; white symbols with +: carrier individuals; white empty symbols: healthy individuals. Left: Family 1 (Fam1); the blue + symbol indicates the presence of the NM_004572.3(PKP2):c.2013delC variant, the orange + symbol indicates the presence of the four Fam1 variants ENSP00000312435.2(DAG1):p.Leu86Phe, ENSP00000263347.7(TCF25):p.Ser390Phe, ENSP00000259371.2(DAB2IP):p.Asp10Gly, ENSP00000357816.5(CTBP2):p.Gly70Arg. Right: Family 2 (Fam2); the pink + symbol indicates the presence of the NG_009000.1(PKP2):c.2569_2577 + 41del variant, the green + symbol indicates the presence of the ENSP00000434586.1(TTN):p.Gln24857His and the ENSP00000434586.1(TTN):p.Arg23483His variants
