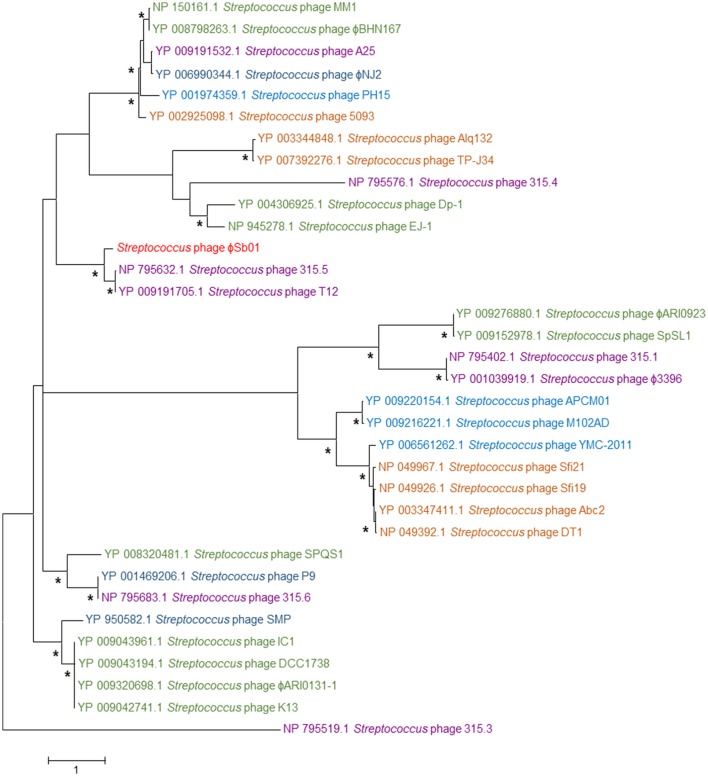
Figure 6.
Maximum likelihood phylogeny of the phage terminase large subunit (TerL) proteins of the Streptococcus phage ϕSb01 and representative reference phages infecting the genus Streptococcus. The tree with the highest log likelihood (−21,218.68) based on the Whelan and Goldman model (Whelan and Goldman, 2001) with frequencies and a gamma distribution with five categories is shown. This tree used a total of 667 amino acid positions and is drawn to scale, with branch lengths measured in the number of substitutions per site and is rooted with the TerL protein sequence of the Streptococcus phage 315.3. Bootstrap supports greater than 85%, based on 1,000 replicates, are indicated (*). The TerL proteins are colored according to phage host species and environmental isolation source S. bovis/equinus 2B (rumen, gut-associated); S. pneumoniae (human respiratory tract); S. pyogenes, GAS strains and S. dysgalactiae subsp. equisimilis (human, skin flora, respiratory tract); S. suis and S. equi (non-human, mammal respiratory tract); S. mutans, S. salivarius, and S. gordonii (human dental and oral); S. thermophilus (dairy fermentations, whey).