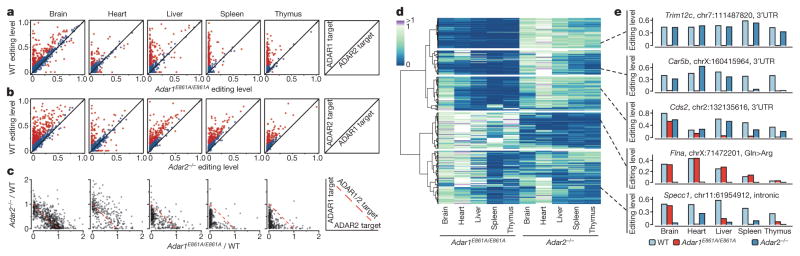
Figure 3. Dynamic regulation of RNA editing by ADAR1 and ADAR2.
a, b, Comparison of editing levels in five tissues between wild-type (WT) and Adar1E861A/E861AIfih1−/− adult mice (a), and between wild-type and Adar2−/−Gria2R/R adult mice (b). Sites that are differentially edited (≥10% editing level difference between wild-type and mutant mouse) are marked in red. The diagram on the right illustrates how ADAR1 and ADAR2 targets are determined by comparing their editing levels between wild-type and mutant mice. c, Comparison of editing ratios between Adar1E861A/E861AIfih1−/− and wild-type mice, and between Adar2−/−Gria2R/R and wild-type mice. The editing ratio is defined as the editing level in the ADAR editing-deficient mutant divided by the editing level in the wild-type mouse. The diagram on the right illustrates how ADAR1 and ADAR2 targets are determined by comparing their editing ratios. d, Heat map and dendrogram of hierarchical clustering of editing sites using editing ratios between either Adar1E861A/E861AIfih1−/− or Adar2−/−Gria2R/R and wild-type mice in five adult mouse tissues. The editing sites could be divided into five distinct clusters (1 to 5, top to bottom), three of which exhibit tissue-specific regulation by ADAR1 and ADAR2. Cluster 1, sites mostly affected by ADAR1 but unaffected by ADAR2 in all five tissues; cluster 2, sites mostly affected by ADAR1 in all five tissues while also being affected by ADAR2 in some tissues; cluster 3, sites mostly affected by both ADAR1 and ADAR2 in brain, while mostly by ADAR1 in the other four tissues; cluster 4, sites mostly affected by ADAR2 in all five tissues; cluster 5, sites affected by both ADAR1 and ADAR2 at different levels in five tissues. e, Exemplary sites for each cluster highlighting the complex regulation of A-to-I editing by the ADAR enzymes.